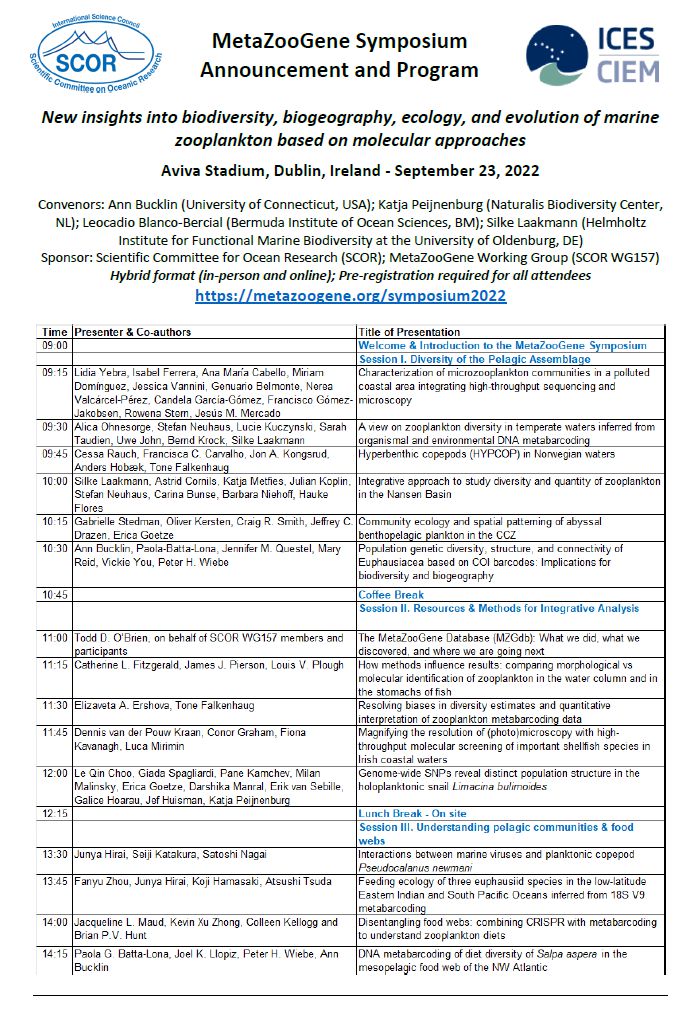
SCOR MetaZooGene Symposium 2022
New insights into biodiversity, biogeography, ecology, and evolution of
marine zooplankton based on molecular approaches
Convenors:
|
Ann Bucklin, University of Connecticut (USA)
Katja Peijnenburg, Naturalis Biodiversity Center and University of Amsterdam (Netherlands)
Leocadio Blanco-Bercial, Bermuda Institute of Ocean Sciences (Bermuda)
Silke Laakmann, Helmholtz Institute for Functional Marine Biodiversity at the University of Oldenburg and
Alfred Wegener Institute Helmholtz Center for Polar and Marine Research (Germany)
|
Sponsor: Scientific Committee for Ocean Research (SCOR); MetaZooGene Working Group (SCOR WG157)
Aviva Stadium, Dublin, Ireland September 23, 2022
Marine zooplankton are key players in pelagic food webs, central links in ecosystem function, useful indicators of water masses, and rapid responders to environmental variation and climate change. This symposium seeks to explore advances in molecular approaches to analysis of biodiversity, biogeography, ecology, and evolution of marine zooplankton, with particular focus on integration with other analytical and observational approaches, including morphological microscopic taxonomic identification, optical and acoustic imagery, trophic biomarker analysis, and more. Discussion provided new insights into the promise and prospects of molecular analysis and recent methodological advancements for accurate identification of species, quantitative (abundance and biomass) measurement, analysis of trophic relationships and food web structure, and more. This symposium provided a platform for participants to network and discuss interdisciplinary and innovative approaches to future zooplankton research.
Below are selected presentations from the SCOR MetaZooGene Symposium,
shared with permission of the presenting authors.
Presentation (Audio/Video)
Batta-Lona, Paola (et al): DNA metabarcoding of diet diversity of Salpa aspera in the mesopelagic food web of the NW Atlantic
Koh, Marc (et al): First barcoding of copepod Monstrilloids from two locations in Australian coastal waters (Coral Bay WA and Southern Great Barrier Reef
Lavrador, Ana (et al): Detection and monitoring of non-indigenous invertebrate species in recreational marinas through DNA metabarcoding of zooplankton communities in the North of Portugal
Maathuis, Margot (et al): Development of zooplankton monitoring in the shallow and turbid Dutch Wadden Sea integrating DNA metabarcoding and automatic image analysis
OBrien, Todd (et al): The MetaZooGene Database (MZGdb): What we did, what we discovered, and where we are going next
Rauch, Cessa (et al): g. Hyperbenthic Copepods (HYPCOP) in Norwegian waters
van der Pouw Kraan, Dennis (et al): Magnifying the resolution of (photo)microscopy with high-throughput molecular screening of important shellfish species in Irish coastal waters
Presentation (PDF Slides)
Bucklin, Ann (et al): Population genetic diversity, structure, and connectivity of Euphausiacea based on COI barcodes: Implications for biodiversity and biogeography
Fitzgerald, Catherine (et al): How methods influence results: comparing morphological vs molecular identification of zooplankton in the water column and in the stomachs of fish
Maud, Jacqueline (et al): Disentangling food webs: combining CRISPR with metabarcoding to understand zooplankton diets
Ohnesorge, Alica (et al): A view on zooplankton diversity in temperate waters inferred from organismal and environmental DNA metabarcoding
Yebra, Lidia (et al): Characterization of microzooplankton communities in a polluted coastal area integrating high-throughput sequencing and microscopy
Zhou, Fanyu (et al): Feeding ecology of three euphausiid species in the low-latitude Eastern Indian and South Pacific Oceans inferred from 18S V9 metabarcoding
Presentation (Posters)
Denes, Marcell (et al): Building the Adriatic Calanoid Database with the help of Oxford’s MinION system.
Lavrador, Ana (et al): Detection and monitoring of non-indigenous invertebrate species in recreational marinas through DNA metabarcoding of zooplankton communities in the North of Portugal
|