Taxa Name
| Taxa
Level | Barcode
Data? | Number of
Barcodes
from
any ocean | Number of
Barcodes
from this region | Barcode Locations and
Species Occurence Map | MZGdb Atlas
v2026-m01-01
2026-Jan-24 |
| Amblyops ewingi |
Species
(1) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4042899
R:1:0:0:0 |
| Anchialina grossa |
Species
(2) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
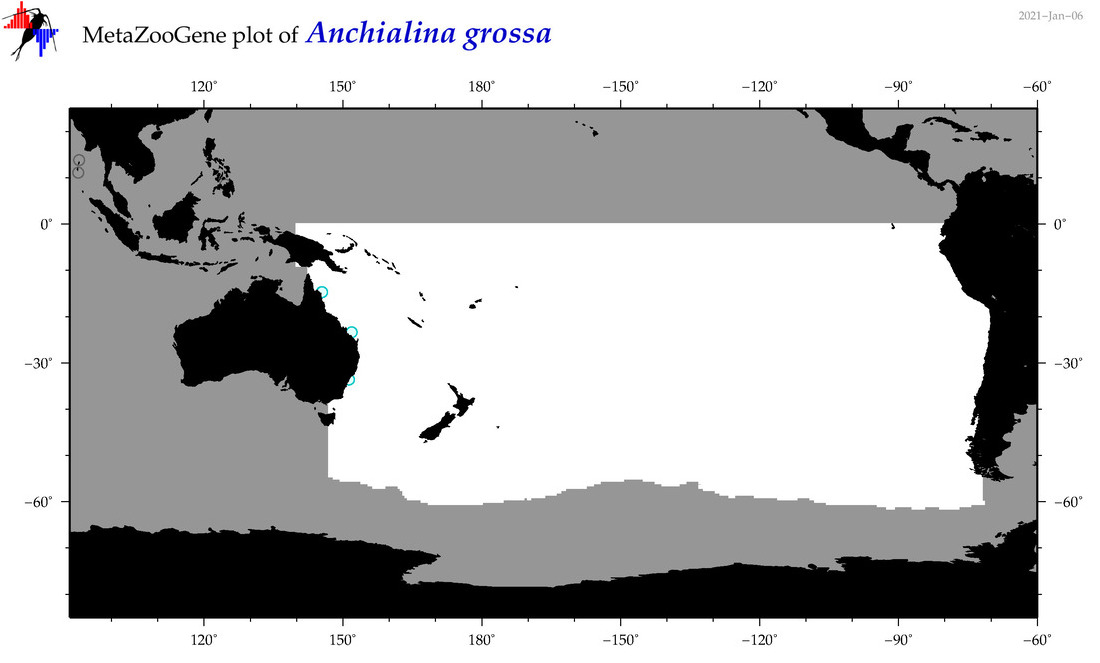 |
accepted
T4043359
R:1:0:0:0 |
| Anchialina penicillata |
Species
(3) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4043364
R:1:0:0:0 |
| Anisomysis (Anisomysis) incisa |
Species
(4) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4094379
R:1:0:0:0 |
| Anisomysis (Anisomysis) laticauda |
Species
(5) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
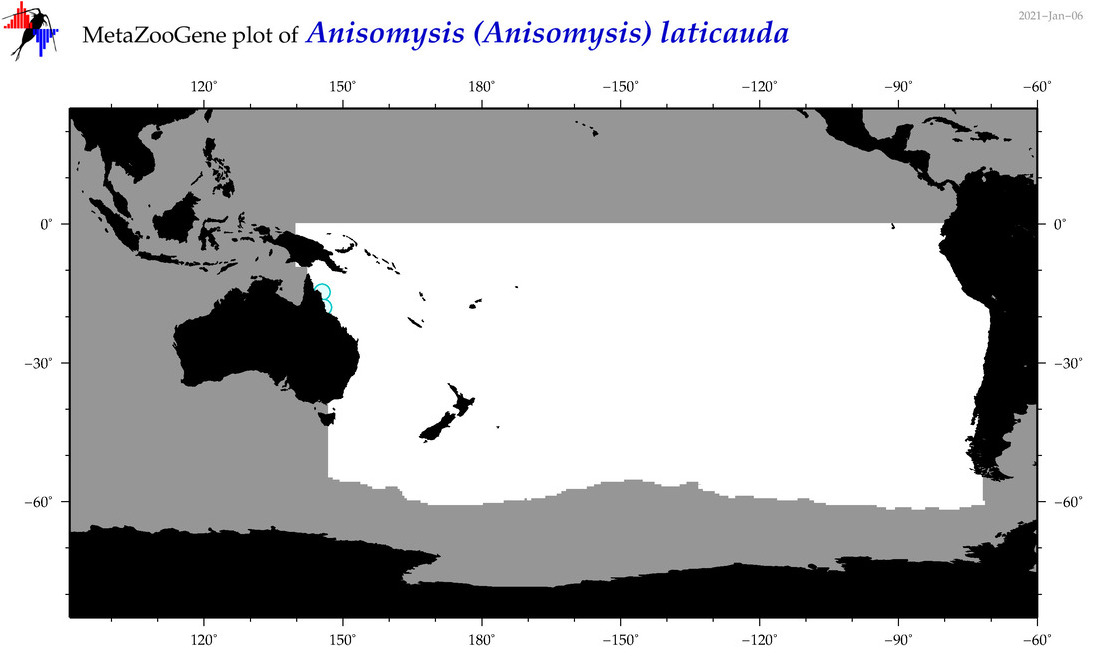 |
accepted
T4094380
R:1:0:0:0 |
| Anisomysis (Anisomysis) parvispina |
Species
(6) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4099986
R:1:0:0:0 |
| Anisomysis (Anisomysis) pelewensis |
Species
(7) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4094528
R:1:0:0:0 |
| Anisomysis (Anisomysis) rotunda |
Species
(8) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4099988
R:1:0:0:0 |
| Anisomysis (Paranisomysis) omorii |
Species
(9) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4099994
R:1:0:0:0 |
| Antarctomysis maxima |
Species
(10) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 0
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
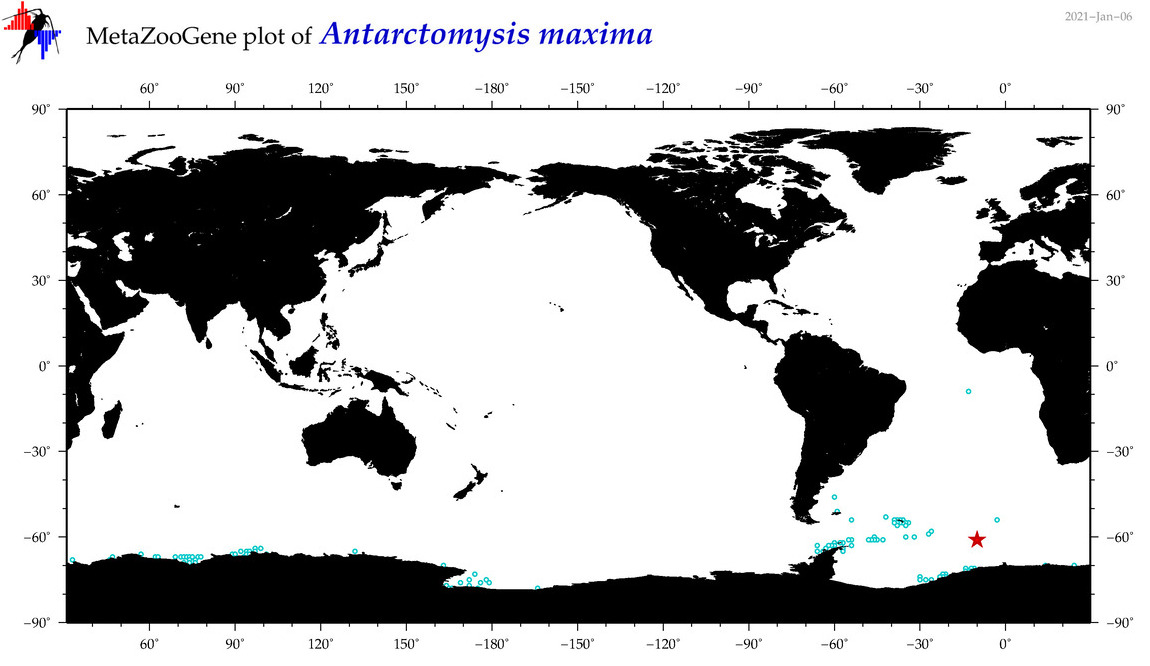 |
accepted
T4007125
R:1:0:0:0 |
| Arachnomysis leuckartii |
Species
(11) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
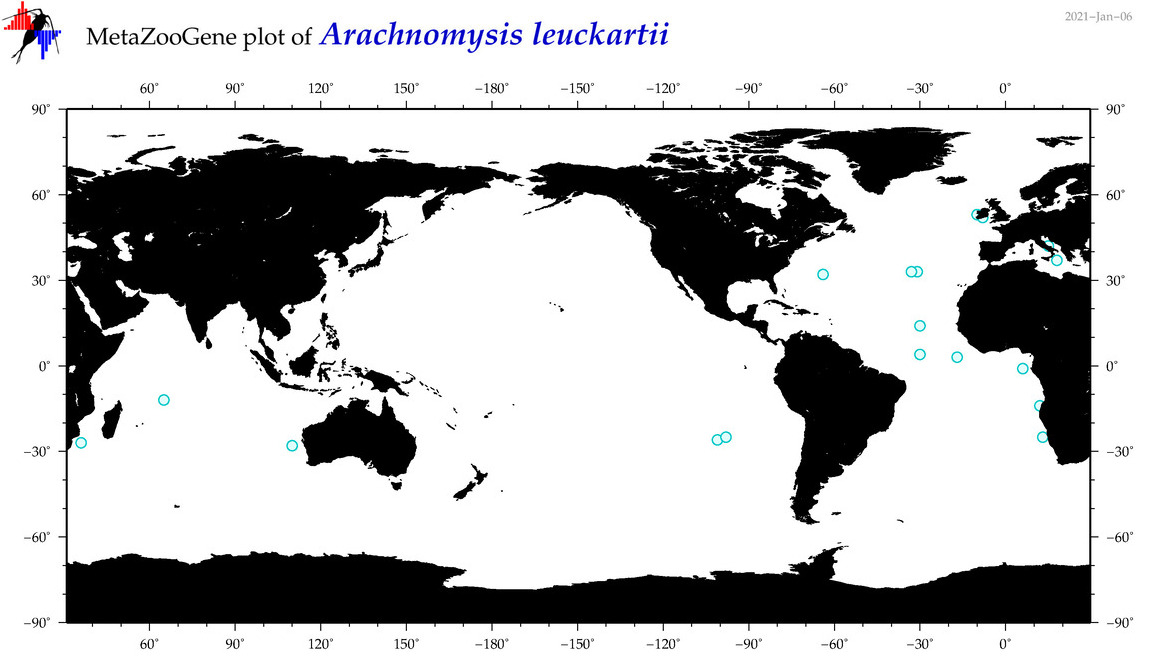 |
accepted
T4044222
R:1:0:0:0 |
| Australomysis acuta |
Species
(12) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
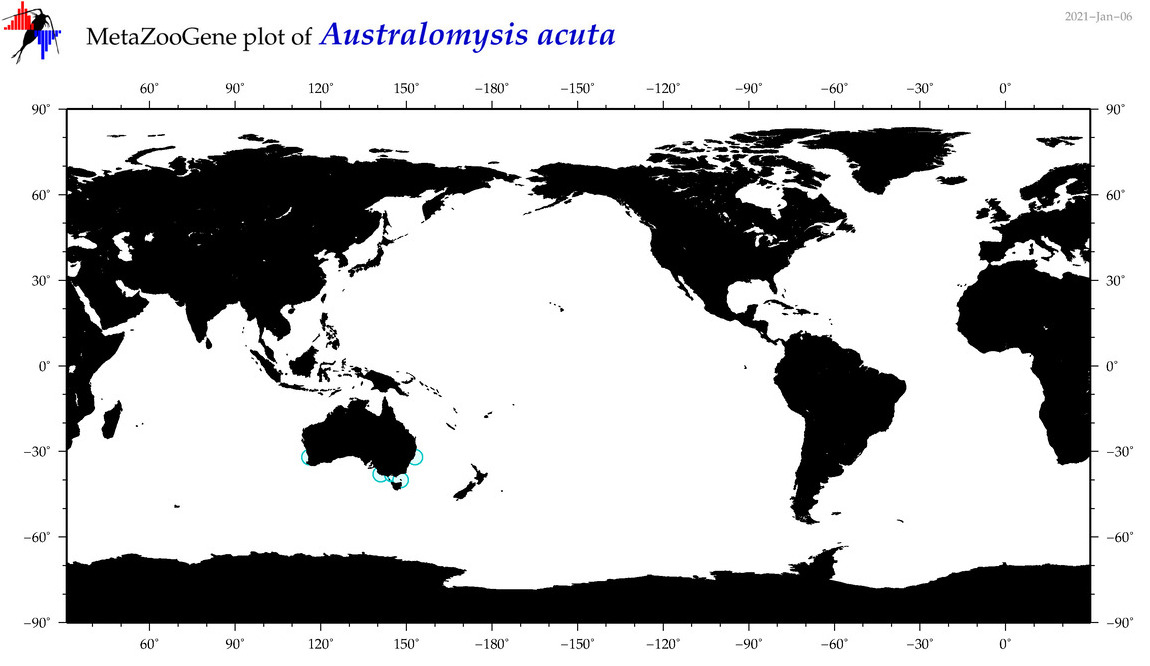 |
accepted
T4044855
R:1:0:0:0 |
| Australomysis aseta |
Species
(13) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4044856
R:1:0:0:0 |
| Australomysis incisa |
Species
(14) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
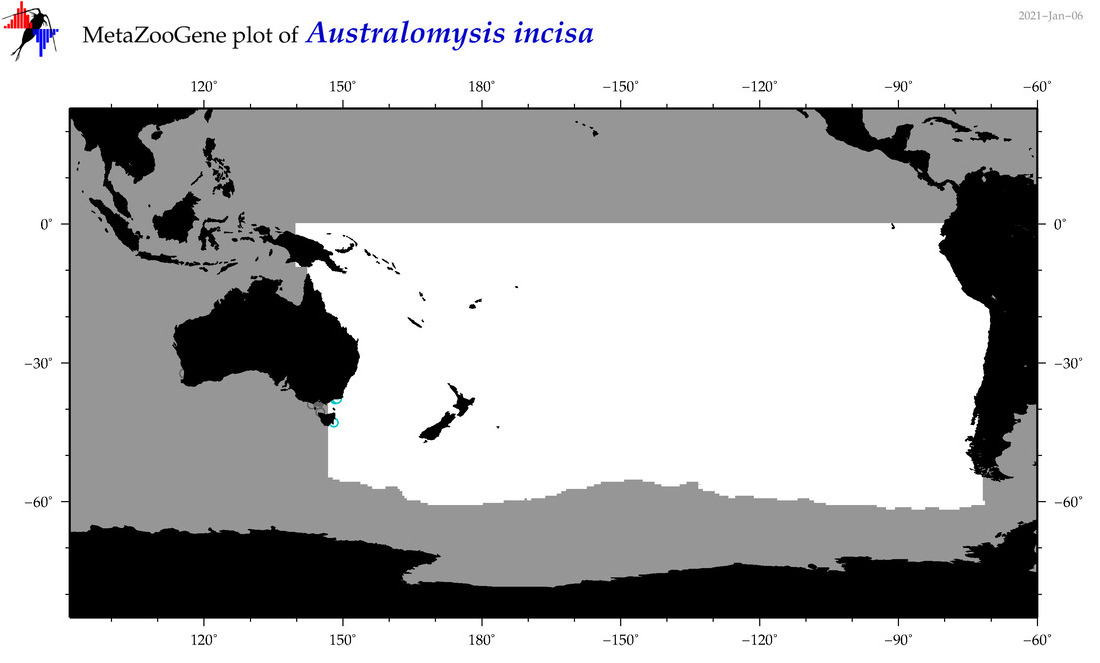 |
accepted
T4007128
R:1:0:0:0 |
| Bacescomysis birsteini |
Species
(15) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
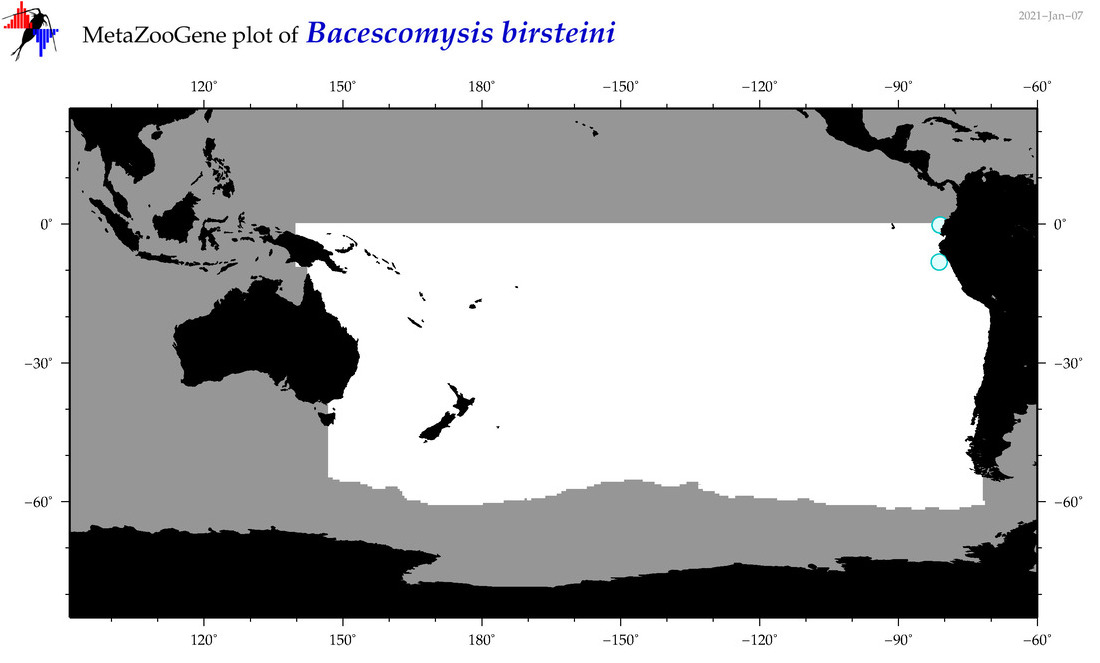 |
accepted
T4045024
R:1:0:0:0 |
| Bacescomysis peruvianus |
Species
(16) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4045026
R:1:0:0:0 |
| Bacescomysis tattersallae |
Species
(17) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4045027
R:1:0:0:0 |
| Boreomysis (Boreomysis) californica |
Species
(18) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
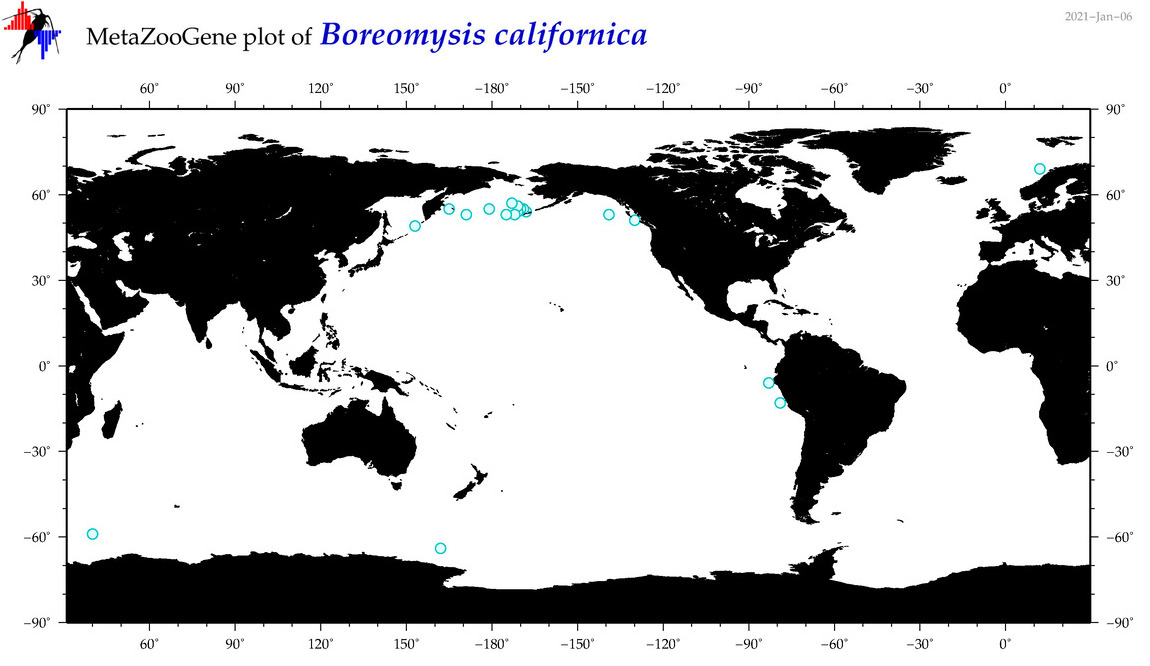 |
accepted
T4045709
R:1:0:0:0 |
| Boreomysis (Boreomysis) fragilis |
Species
(19) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
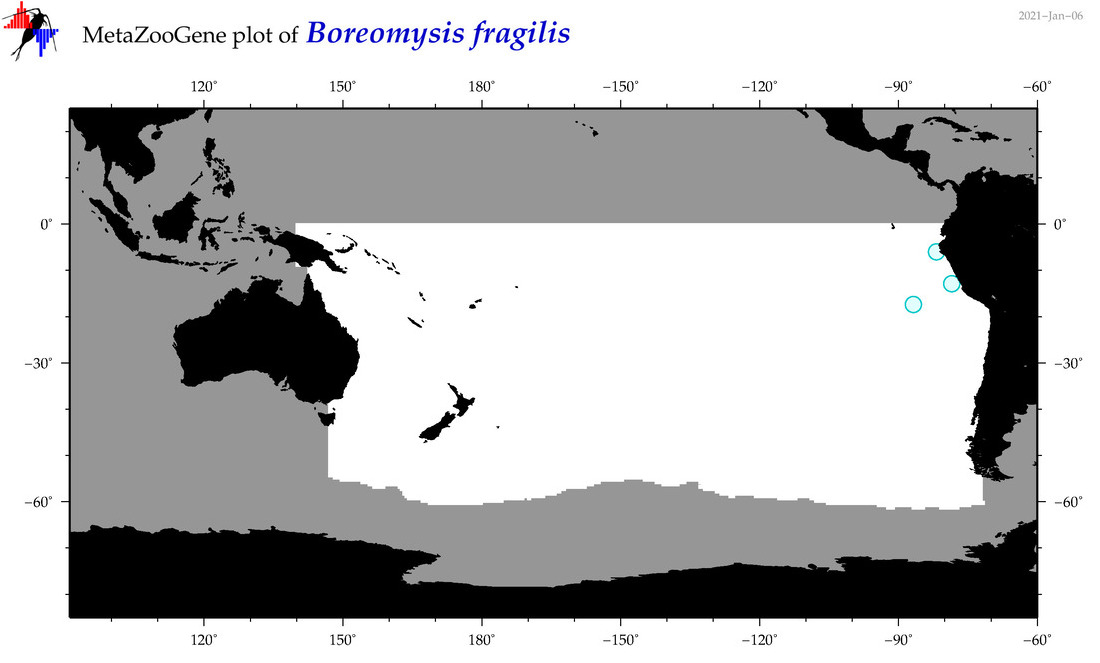 |
accepted
T4045710
R:1:0:0:0 |
| Boreomysis (Boreomysis) rostrata |
Species
(20) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
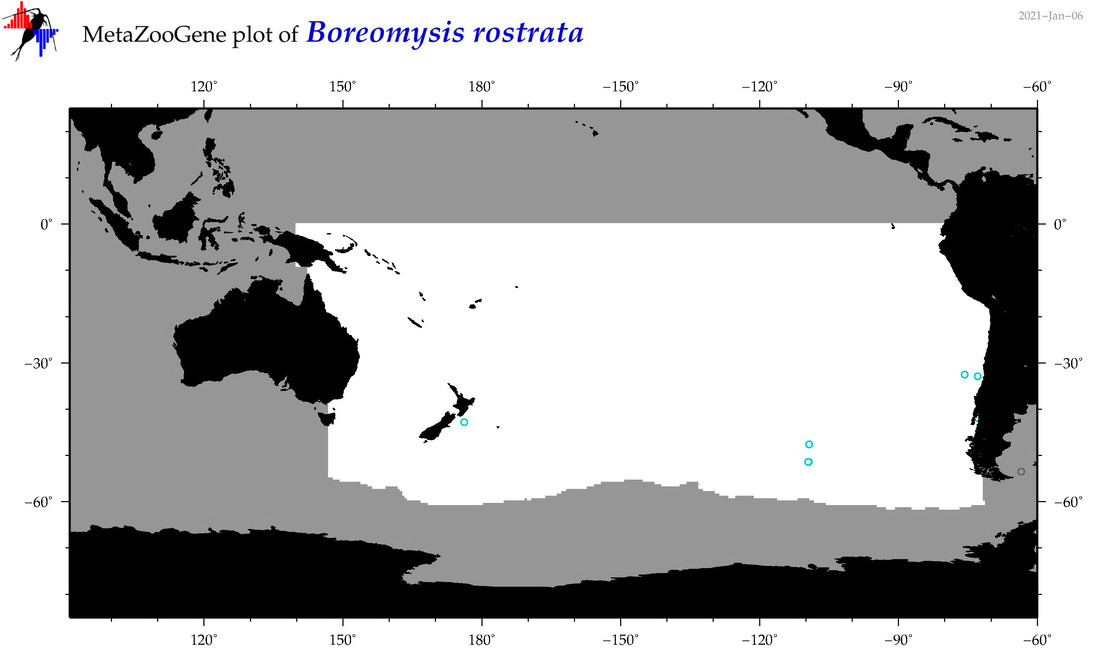 |
accepted
T4007133
R:1:1:0:0 |
| Caesaromysis hispida |
Species
(21) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
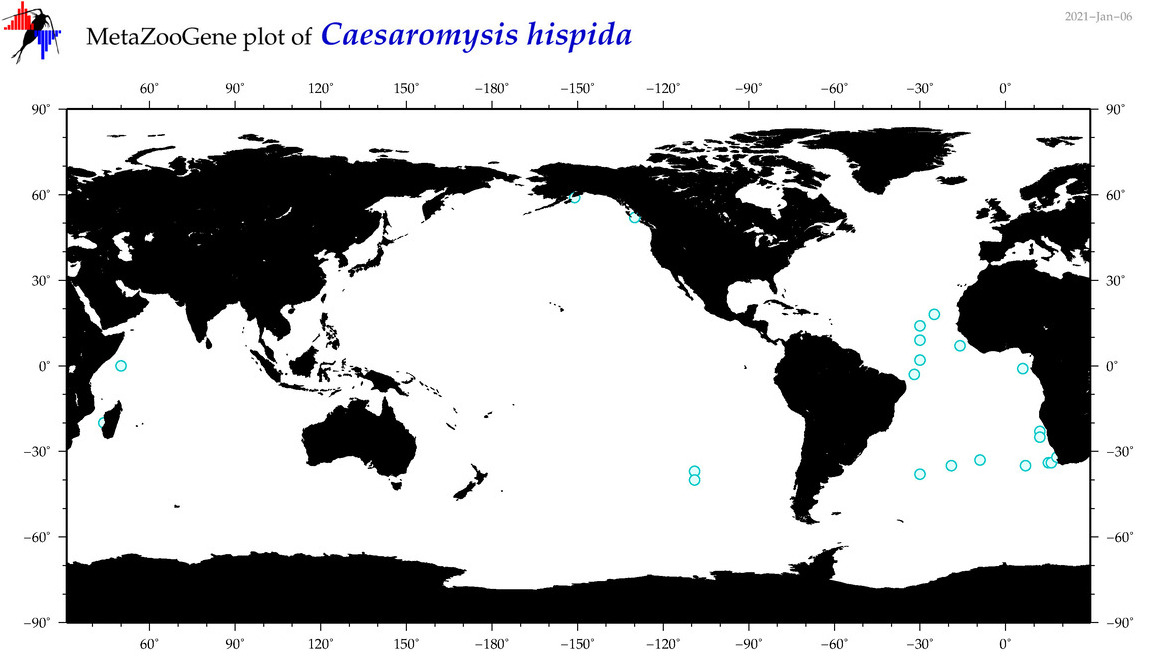 |
accepted
T4003387
R:1:0:0:0 |
| Corellamysis eltanina |
Species
(22) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4048403
R:1:0:0:0 |
| Dactylamblyops fervidus |
Species
(23) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4049212
R:1:0:0:0 |
| Doxomysis acanthina |
Species
(24) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
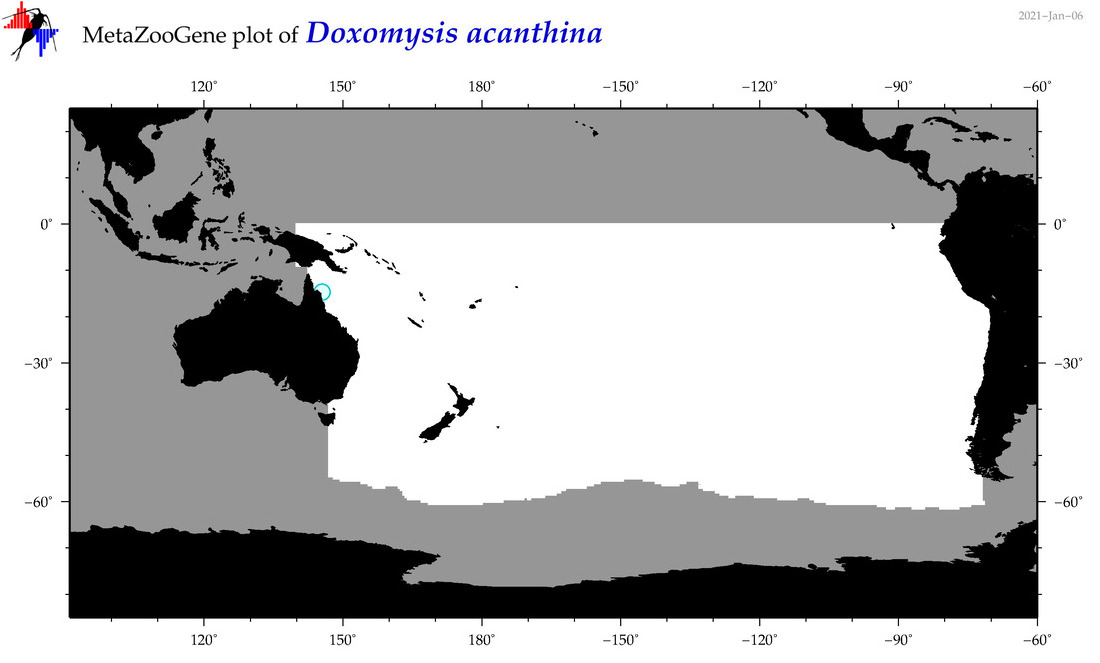 |
accepted
T4050190
R:1:0:0:0 |
| Doxomysis australiensis |
Species
(25) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4050193
R:1:0:0:0 |
| Doxomysis proxima |
Species
(26) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4050198
R:1:0:0:0 |
| Echinomysis serratus |
Species
(27) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
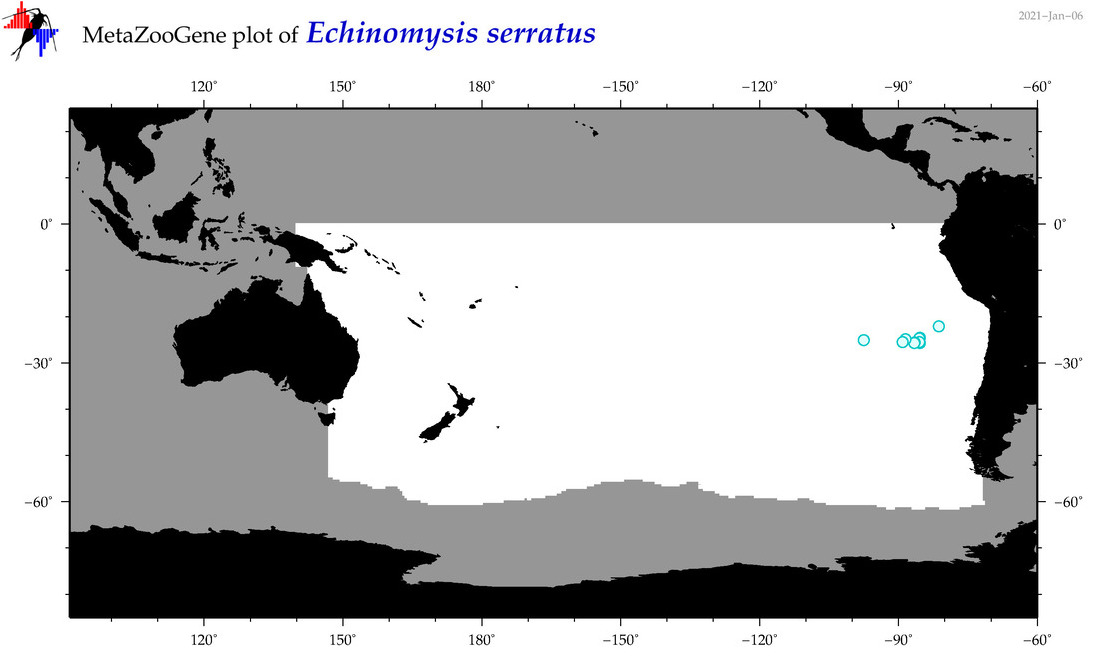 |
accepted
T4050408
R:1:0:0:0 |
| Euchaetomera oculata |
Species
(28) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4051241
R:1:0:0:0 |
| Euchaetomera plebeja |
Species
(29) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4051242
R:1:0:0:0 |
| Euchaetomera tenuis |
Species
(30) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
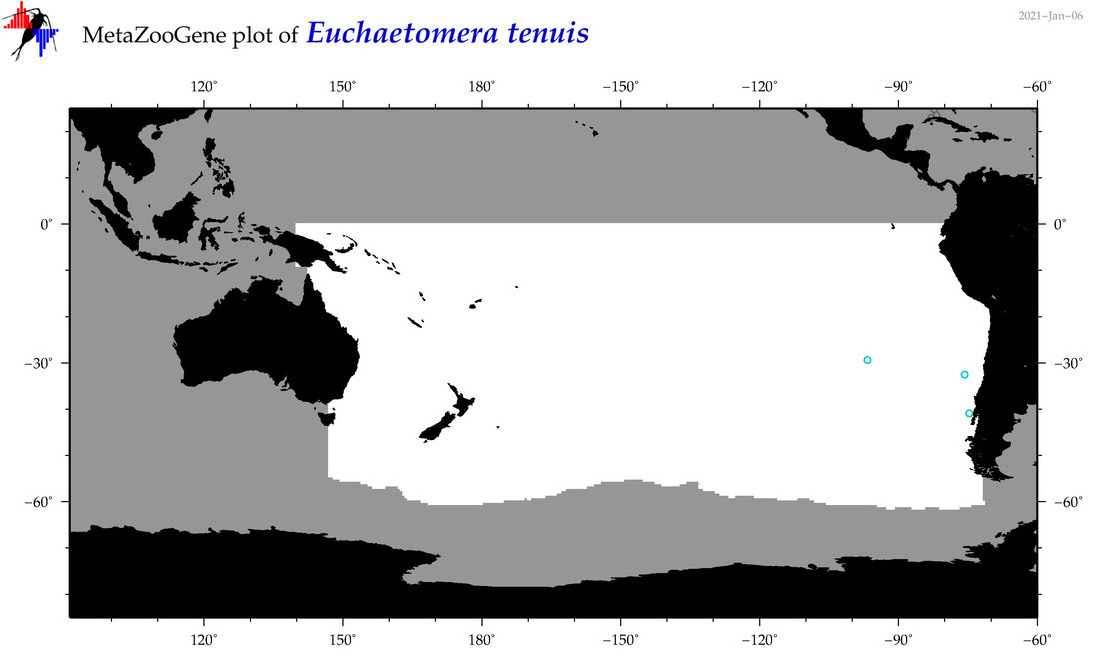 |
accepted
T4007144
R:1:0:0:0 |
| Euchaetomera typica |
Species
(31) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
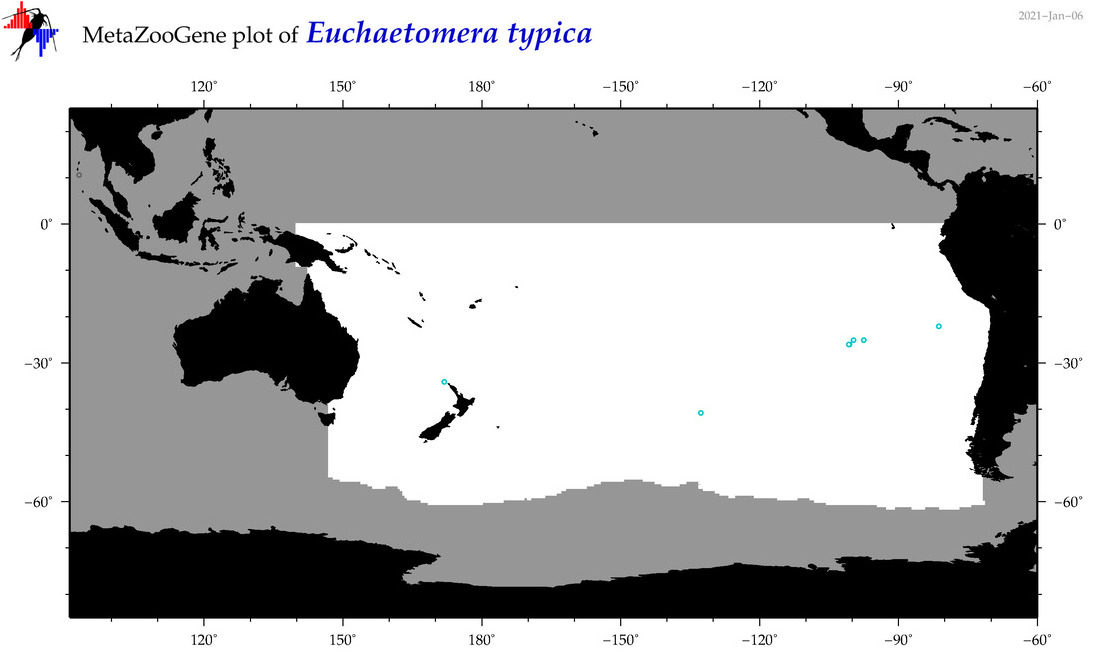 |
accepted
T4007145
R:1:0:0:0 |
| Euchaetomera zurstrasseni |
Species
(32) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
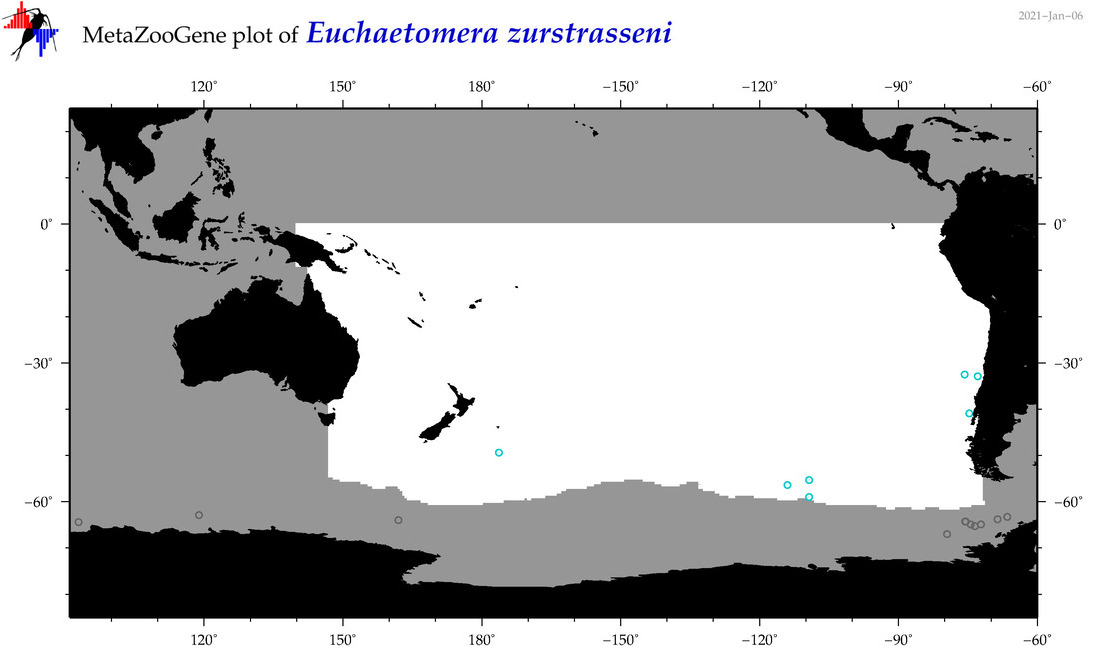 |
accepted
T4007146
R:1:0:0:0 |
| Gastrosaccus australis |
Species
(33) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4099969
R:1:0:0:0 |
| Gastrosaccus daviei |
Species
(34) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4052594
R:1:0:0:0 |
| Hansenomysis carinata |
Species
(35) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4053463
R:1:0:0:0 |
| Hansenomysis chini |
Species
(36) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4053464
R:1:0:0:0 |
| Hansenomysis menziesi |
Species
(37) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
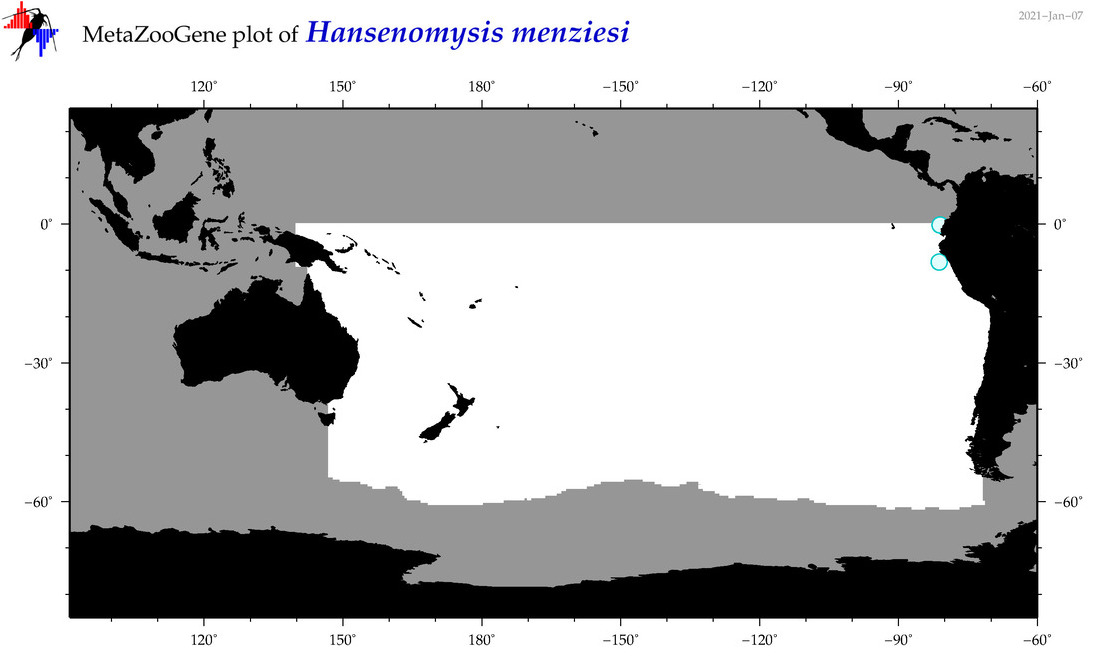 |
accepted
T4053466
R:1:0:0:0 |
| Hansenomysis spenceri |
Species
(38) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
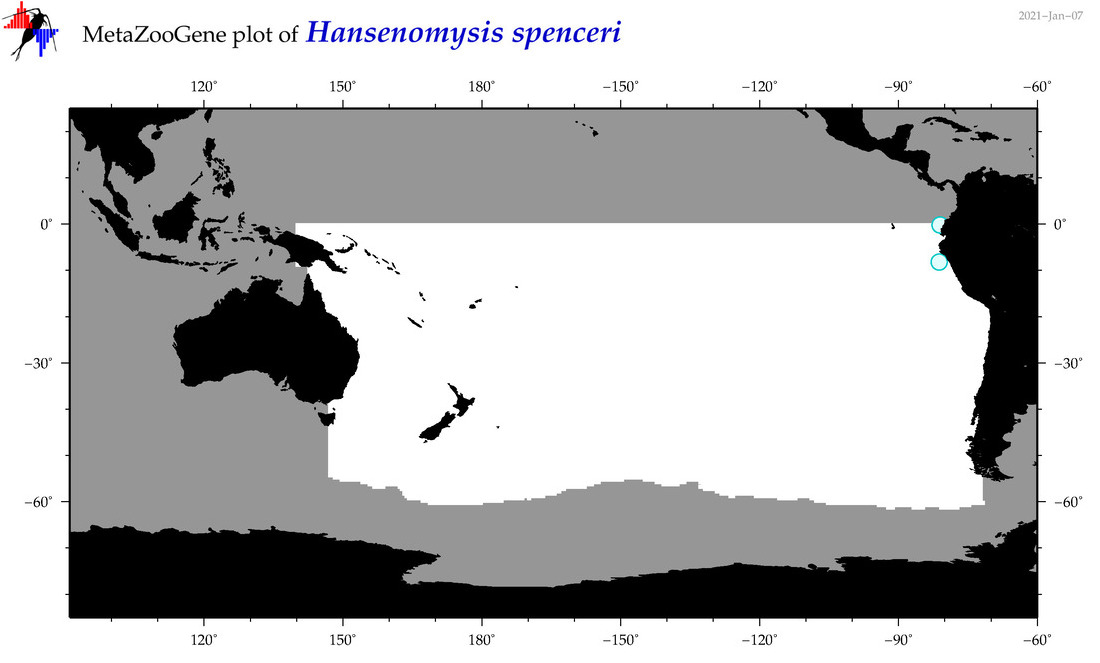 |
accepted
T4053470
R:1:0:0:0 |
| Hansenomysis tropicalis |
Species
(39) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4053471
R:1:0:0:0 |
| Haplostylus bengalensis |
Species
(40) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
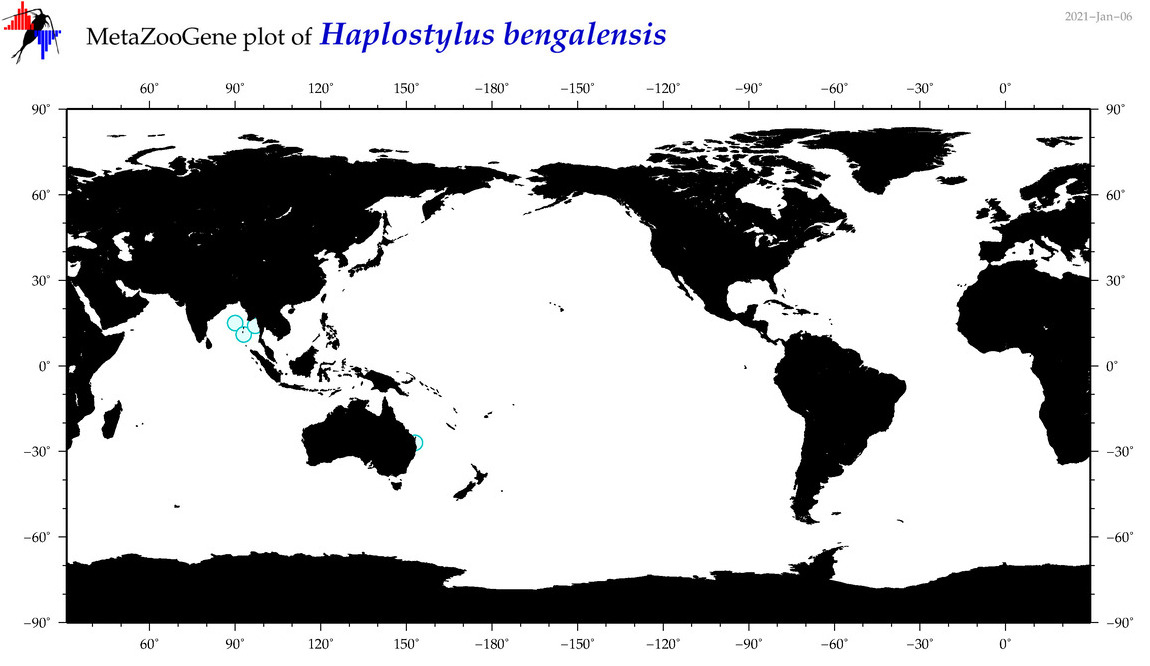 |
accepted
T4053564
R:1:0:0:0 |
| Haplostylus brisbanensis |
Species
(41) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4053565
R:1:0:0:0 |
| Haplostylus indicus |
Species
(42) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
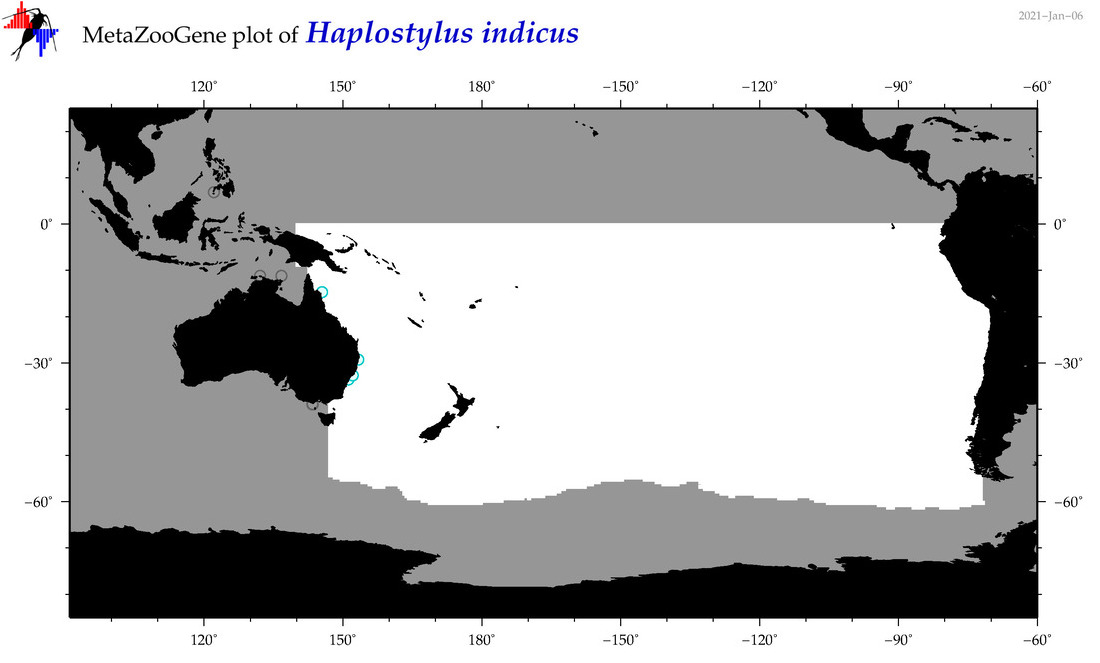 |
accepted
T4053567
R:1:0:0:0 |
| Haplostylus pacificus |
Species
(43) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4053569
R:1:0:0:0 |
| Haplostylus robusta |
Species
(44) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
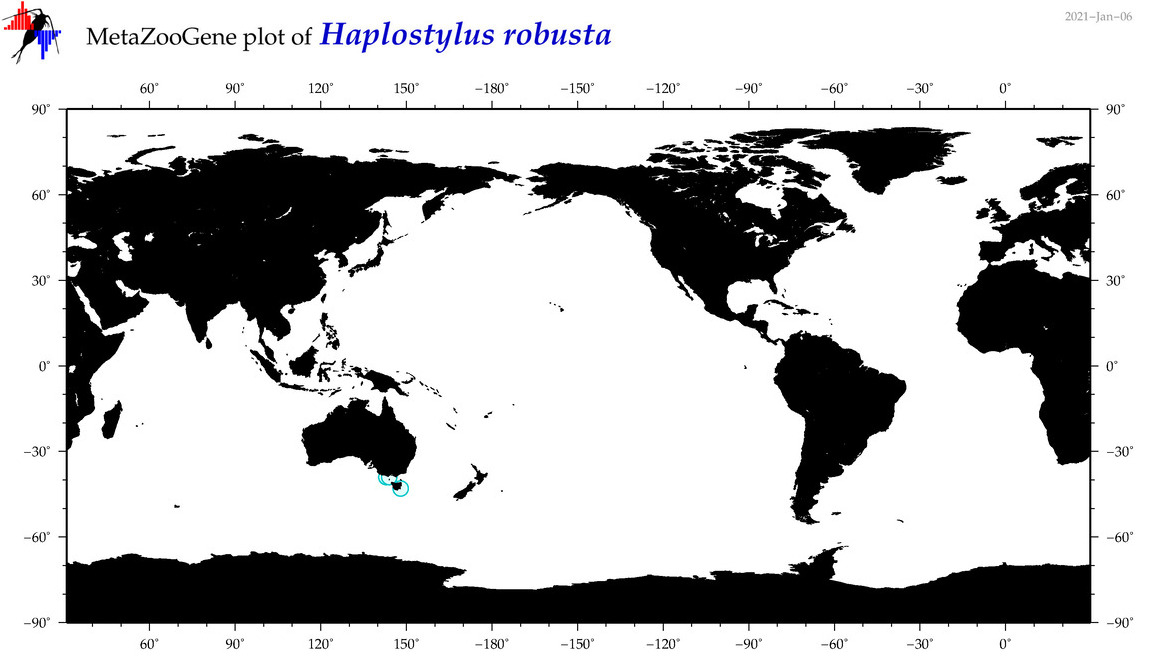 |
accepted
T4053572
R:1:0:0:0 |
| Hemisiriella parva |
Species
(45) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
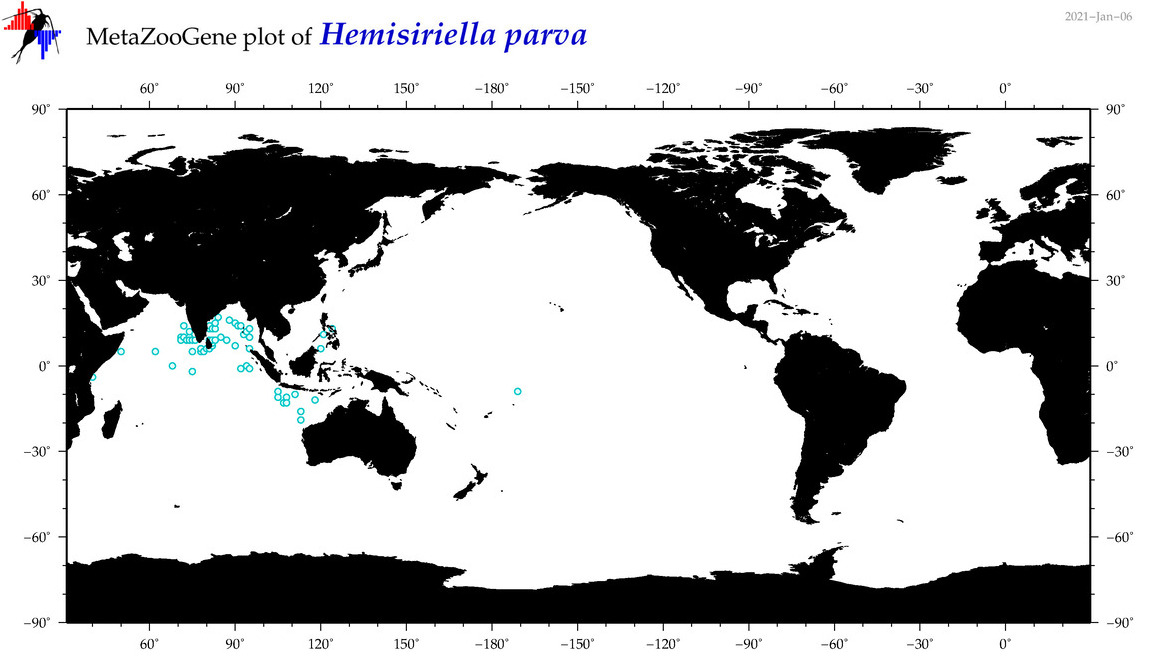 |
accepted
T4007156
R:1:0:0:0 |
| Hemisiriella pulchra |
Species
(46) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
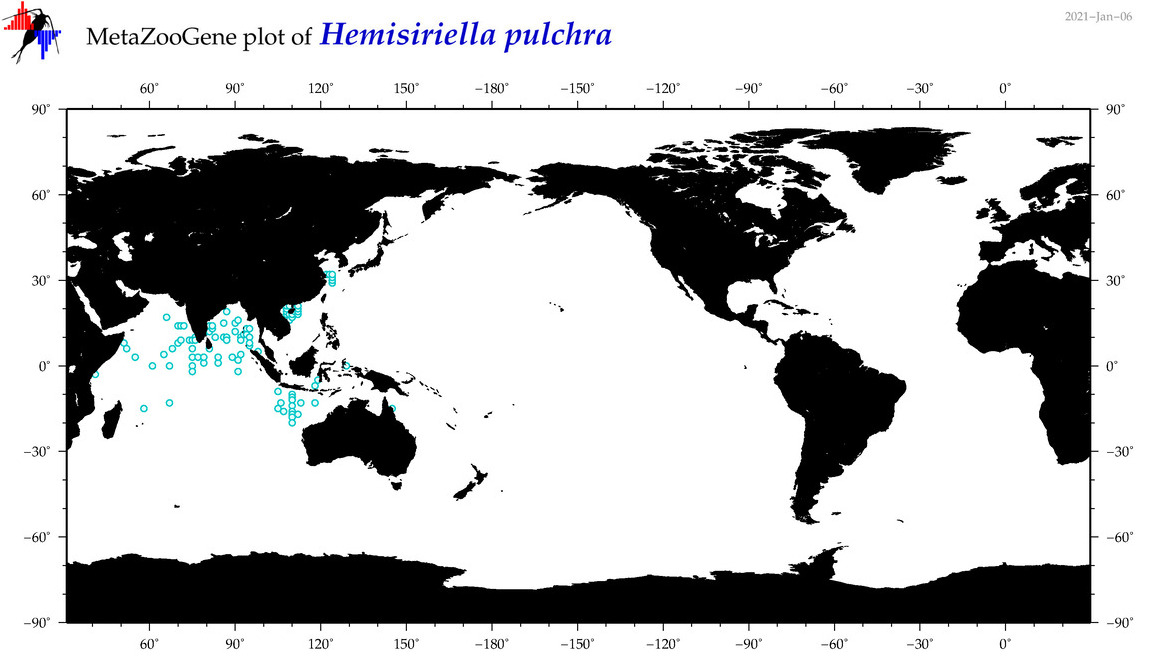 |
accepted
T4007157
R:1:0:0:0 |
| Heteromysis (Gnathomysis) harpax |
Species
(47) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4094522
R:1:0:0:0 |
| Heteromysis (Gnathomysis) harpaxoides |
Species
(48) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
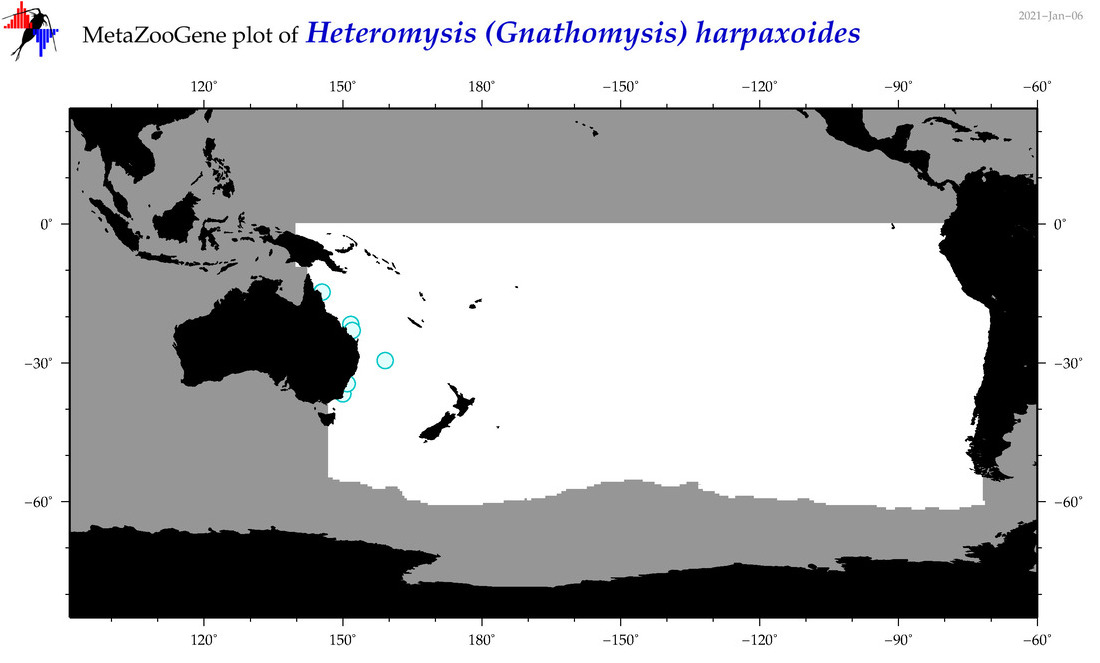 |
accepted
T4094527
R:1:0:0:0 |
| Heteromysis (Heteromysis) australica |
Species
(49) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4094523
R:1:0:0:0 |
| Heteromysis (Heteromysis) communis |
Species
(50) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4094524
R:1:0:0:0 |
| Heteromysis (Heteromysis) tasmanica |
Species
(51) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4099998
R:1:0:0:0 |
| Heteromysis (Heteromysis) tethysiana |
Species
(52) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4099999
R:1:0:0:0 |
| Idiomysis inermis |
Species
(53) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
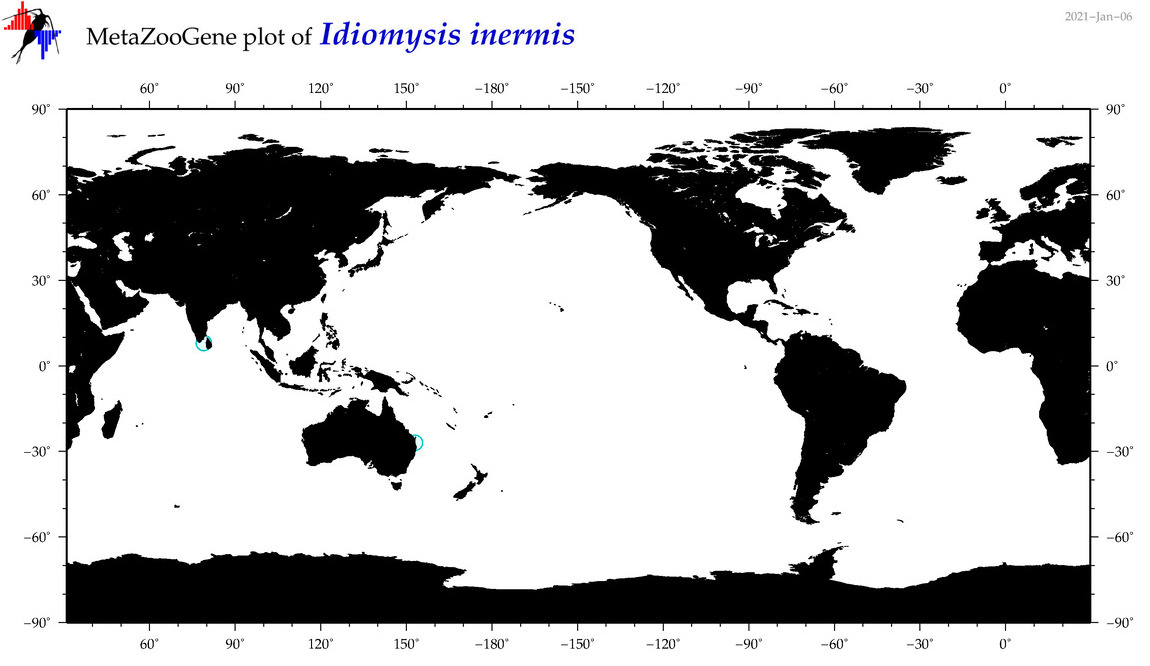 |
accepted
T4054708
R:1:0:0:0 |
| Mysidella australiana |
Species
(54) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
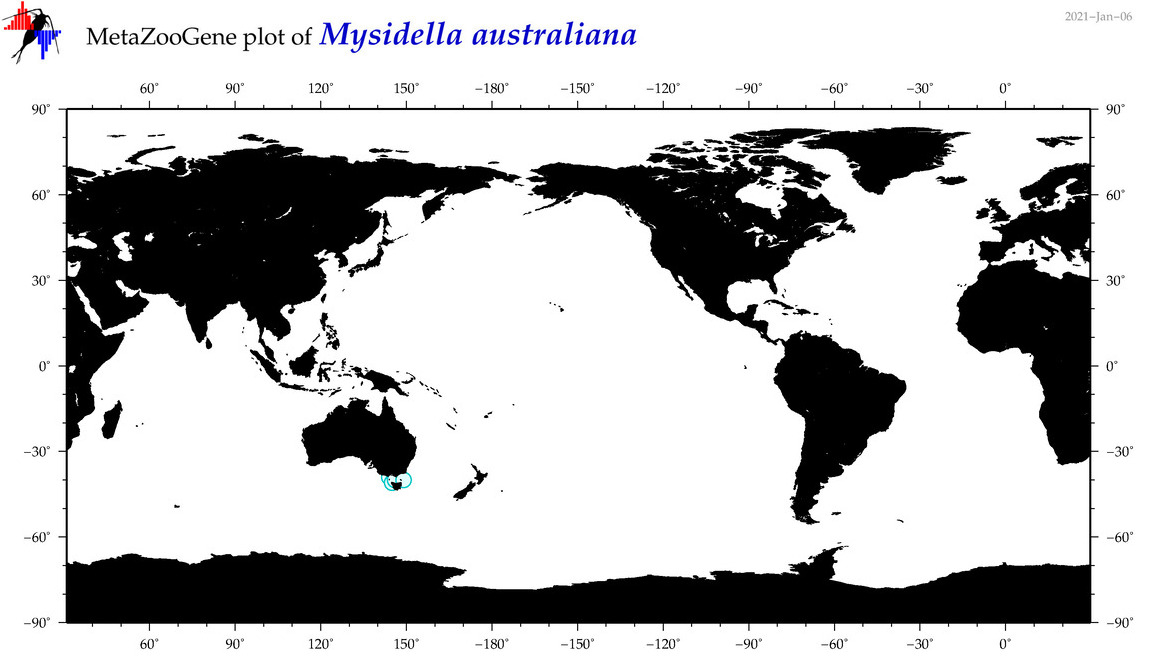 |
accepted
T4059604
R:1:0:0:0 |
| Mysidetes halope |
Species
(55) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4059615
R:1:0:0:0 |
| Mysidetes peruana |
Species
(56) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4059622
R:1:0:0:0 |
| Mysidopsis acuta |
Species
(57) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4007172
R:1:0:0:0 |
| Mysimenzies hadalis |
Species
(58) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4081635
R:1:0:0:0 |
| Neodoxomysis littoralis |
Species
(59) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
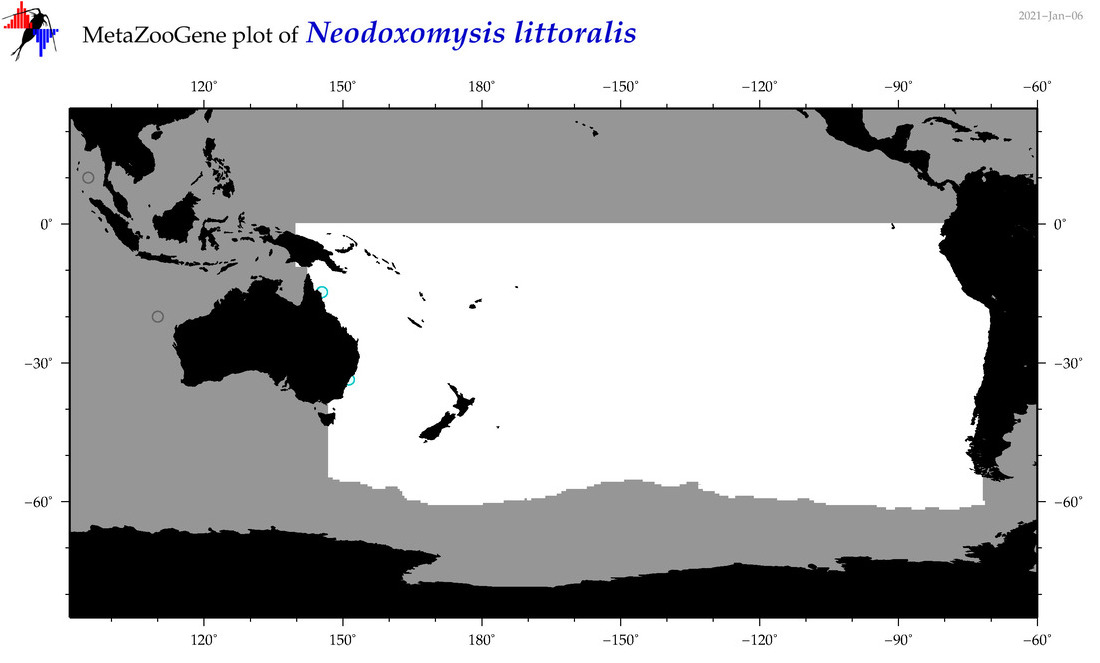 |
accepted
T4060078
R:1:0:0:0 |
| Neomysis ilyapai |
Species
(60) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4060160
R:1:0:0:0 |
| Neomysis japonica |
Species
(61) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 2
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
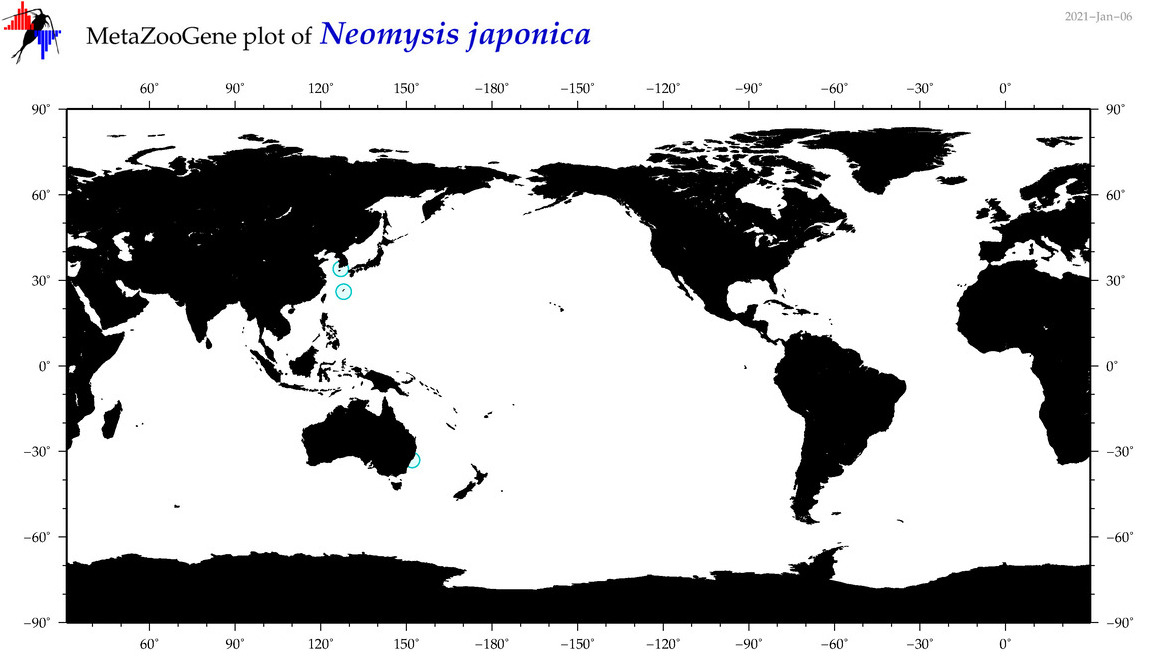 |
accepted
T4003236
R:1:1:0:0 |
| Neomysis sopayi |
Species
(62) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4060167
R:1:0:0:0 |
| Notomysis australiensis |
Species
(63) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4060654
R:1:0:0:0 |
| Paramesopodopsis rufa |
Species
(64) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 1
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
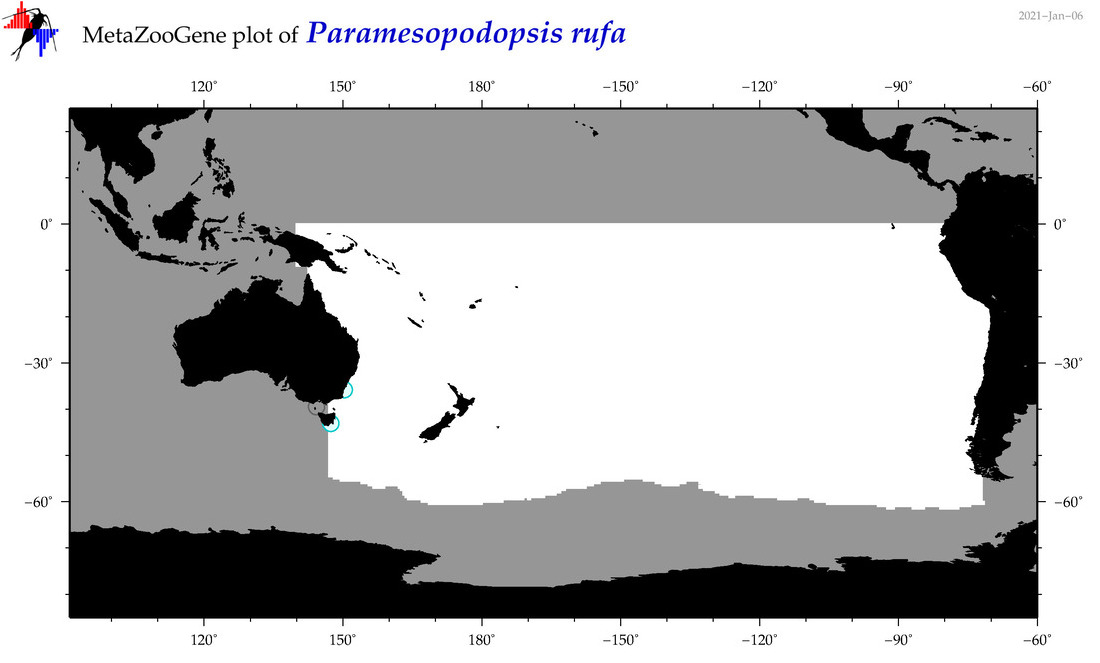 |
accepted
T4013834
R:1:0:0:0 |
| Paranchialina angusta |
Species
(65) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
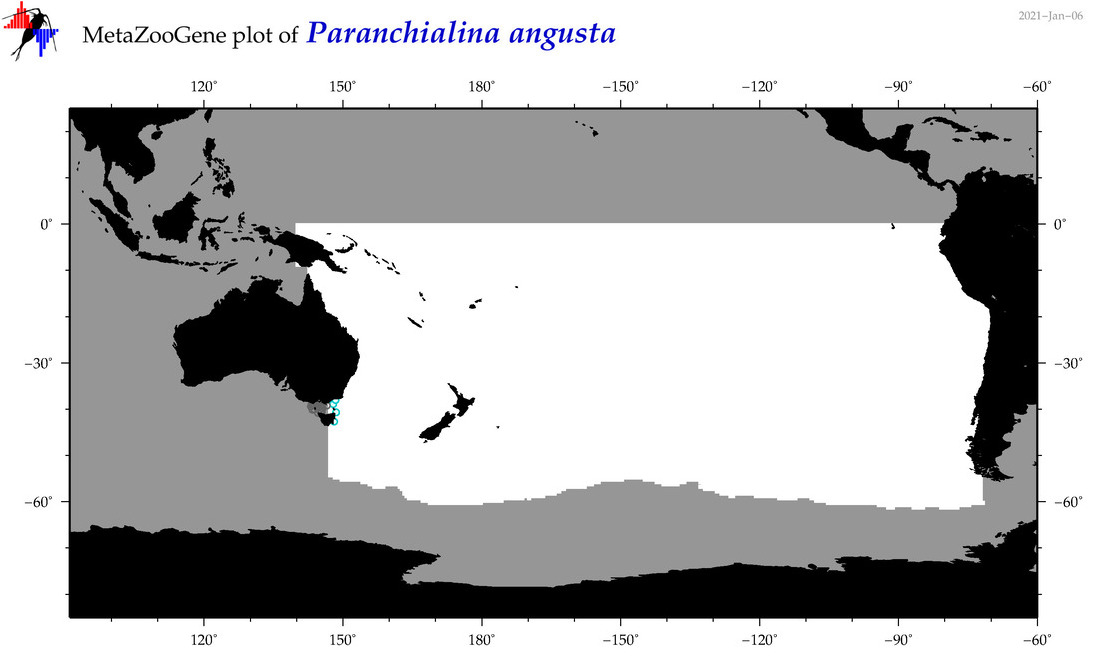 |
accepted
T4007183
R:1:0:0:0 |
| Petalophthalmus armiger |
Species
(66) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
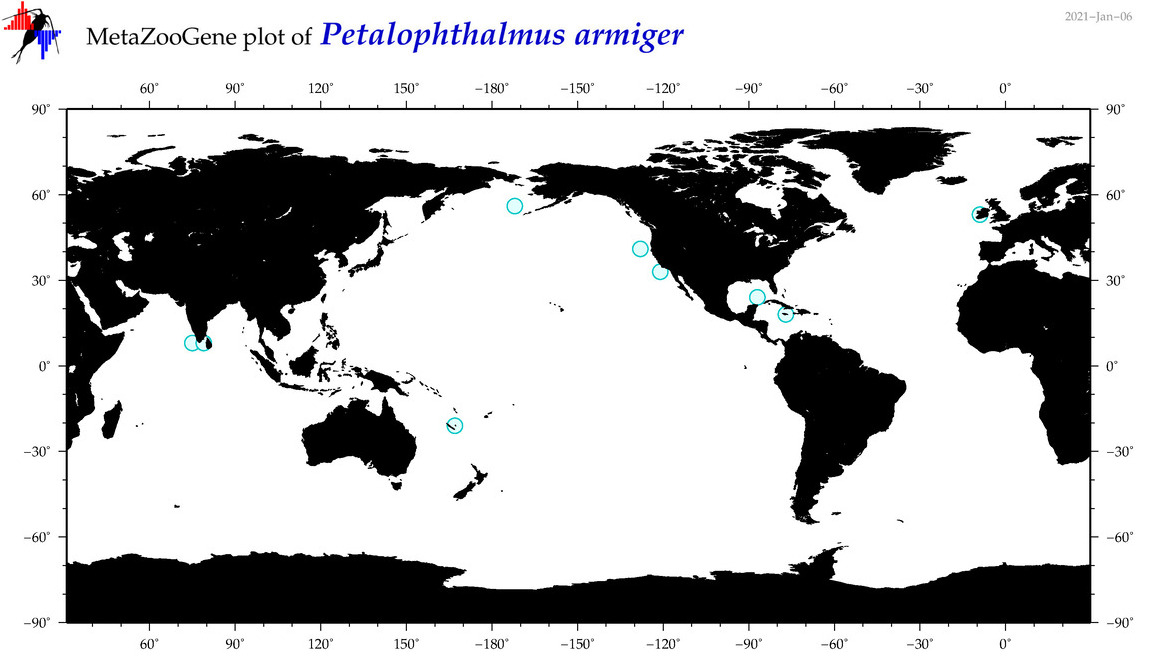 |
accepted
T4063480
R:1:0:0:0 |
| Prionomysis stenolepis |
Species
(67) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
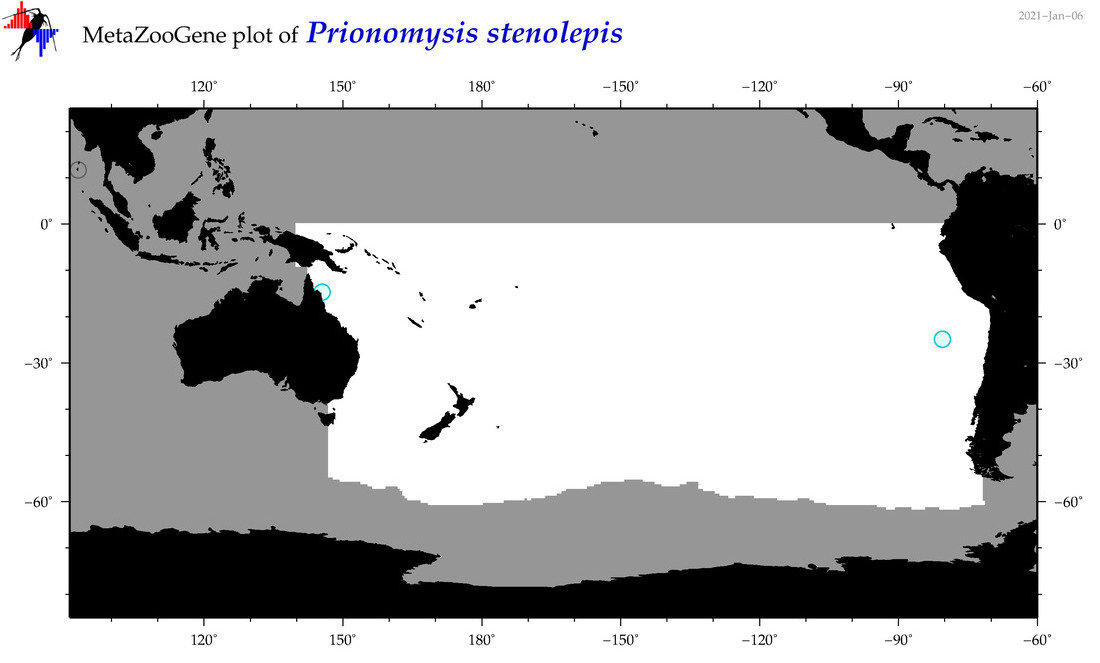 |
accepted
T4065219
R:1:0:0:0 |
| Pseudanchialina inermis |
Species
(68) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
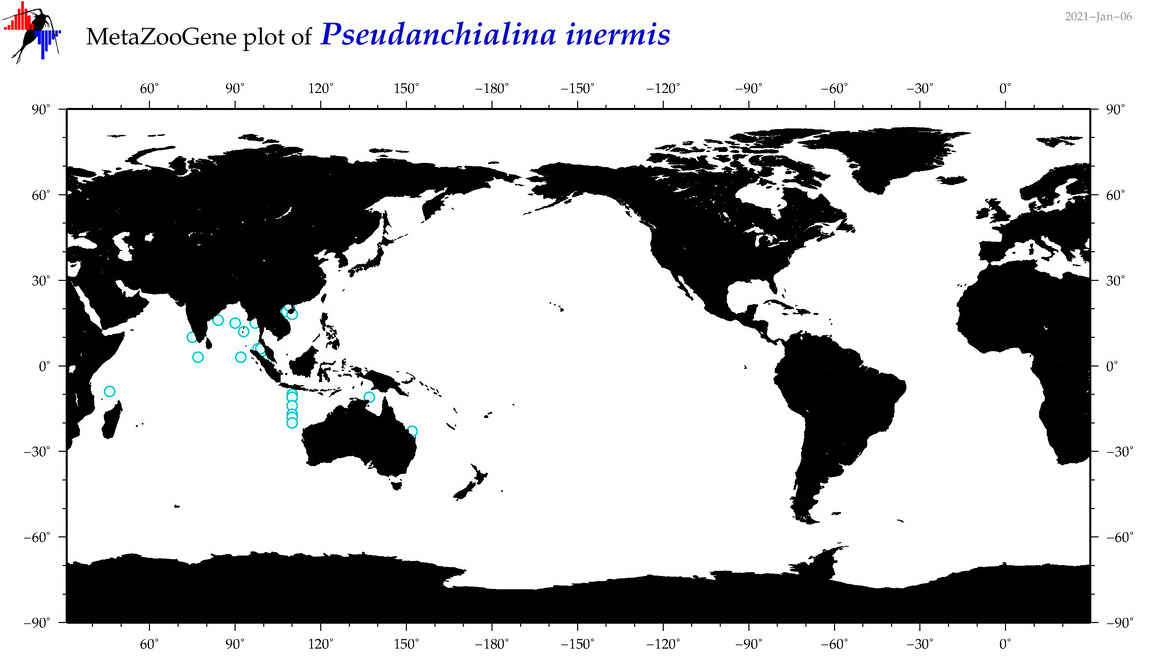 |
accepted
T4007190
R:1:0:0:0 |
| Pseudomma australe |
Species
(69) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
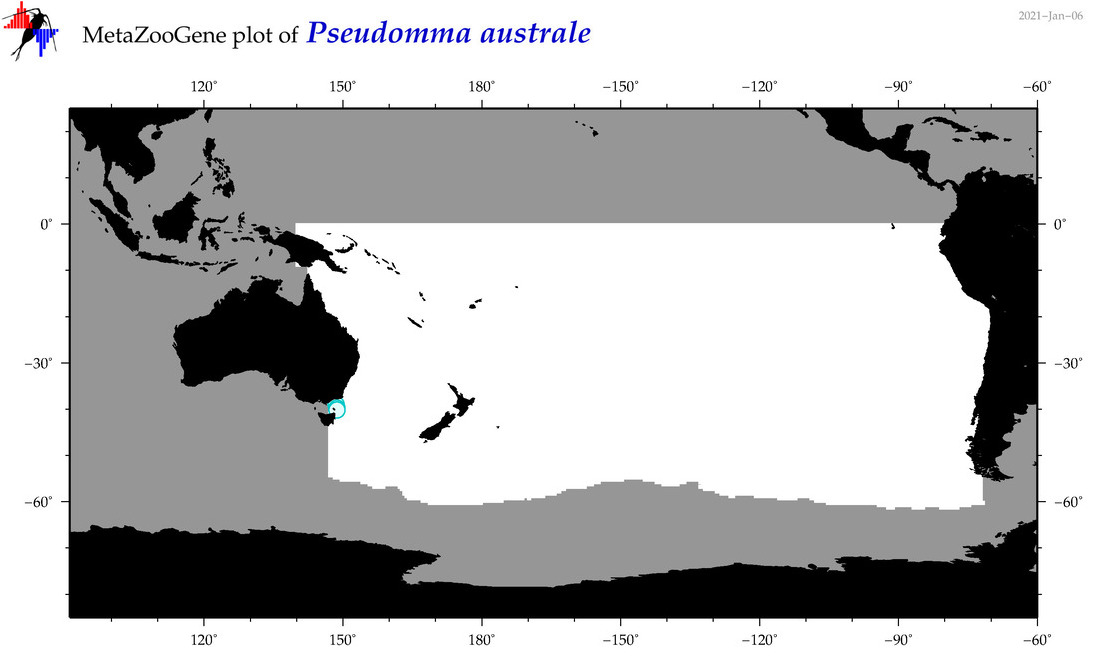 |
accepted
T4066061
R:1:0:0:0 |
| Pseudomma calmani |
Species
(70) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
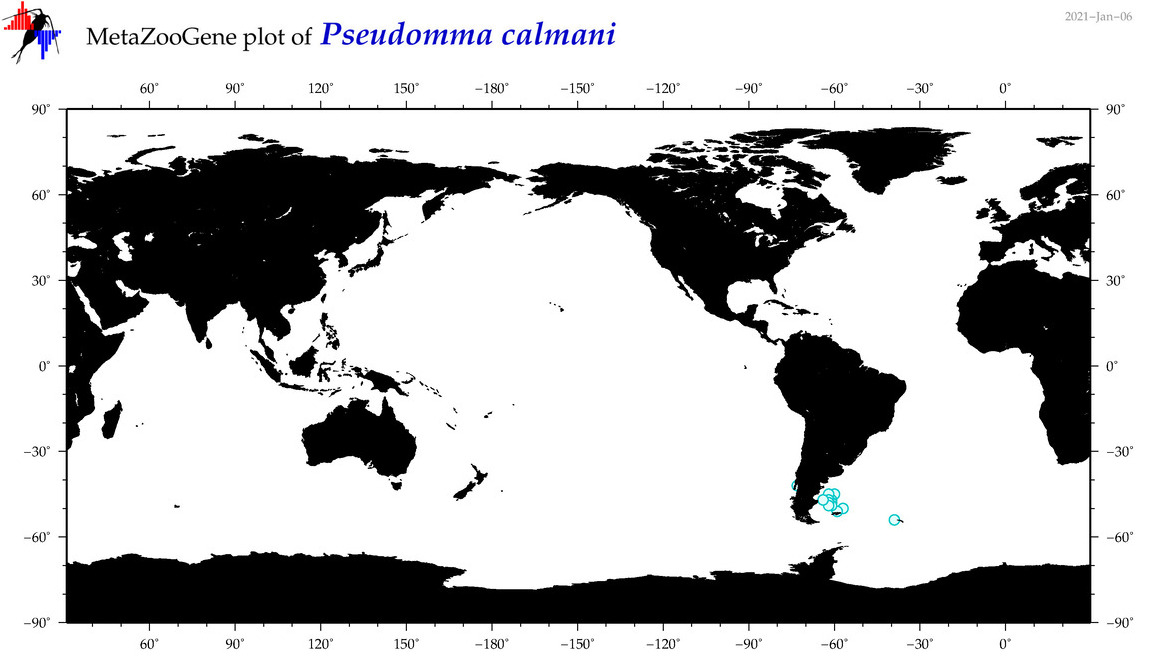 |
accepted
T4066065
R:1:0:0:0 |
| Rostromysis bacescuii |
Species
(71) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4067034
R:1:0:0:0 |
| Siriella affinis |
Species
(72) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
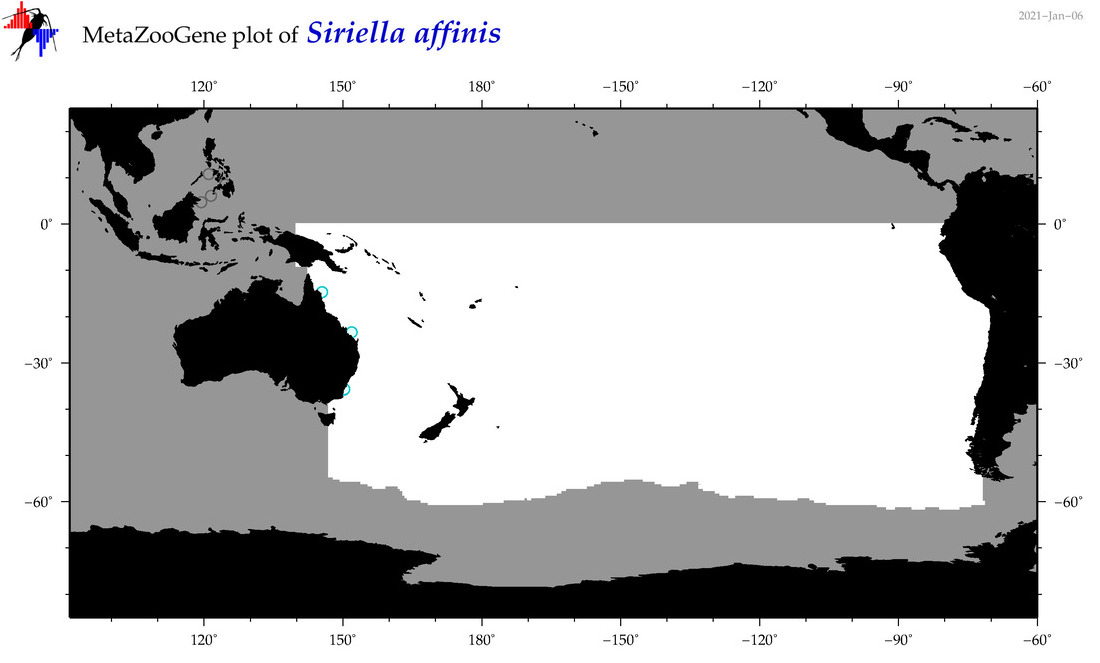 |
accepted
T4068199
R:1:0:0:0 |
| Siriella anomala |
Species
(73) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4068201
R:1:0:0:0 |
| Siriella australis |
Species
(74) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
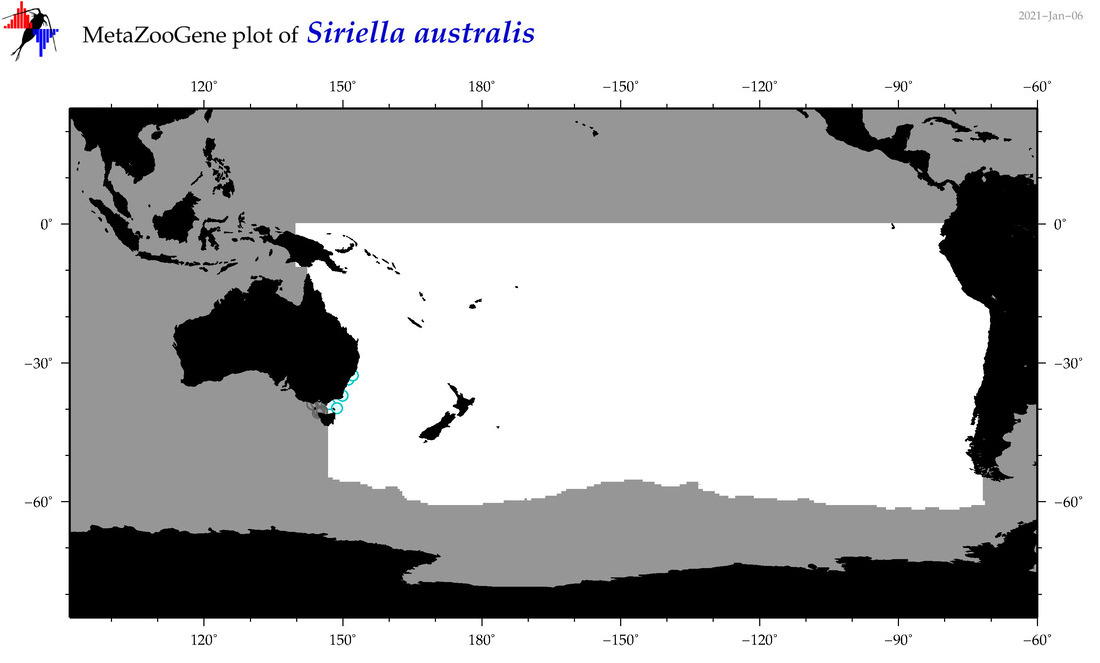 |
accepted
T4068202
R:1:0:0:0 |
| Siriella denticulata |
Species
(75) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4099978
R:1:0:0:0 |
| Siriella distinguenda |
Species
(76) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
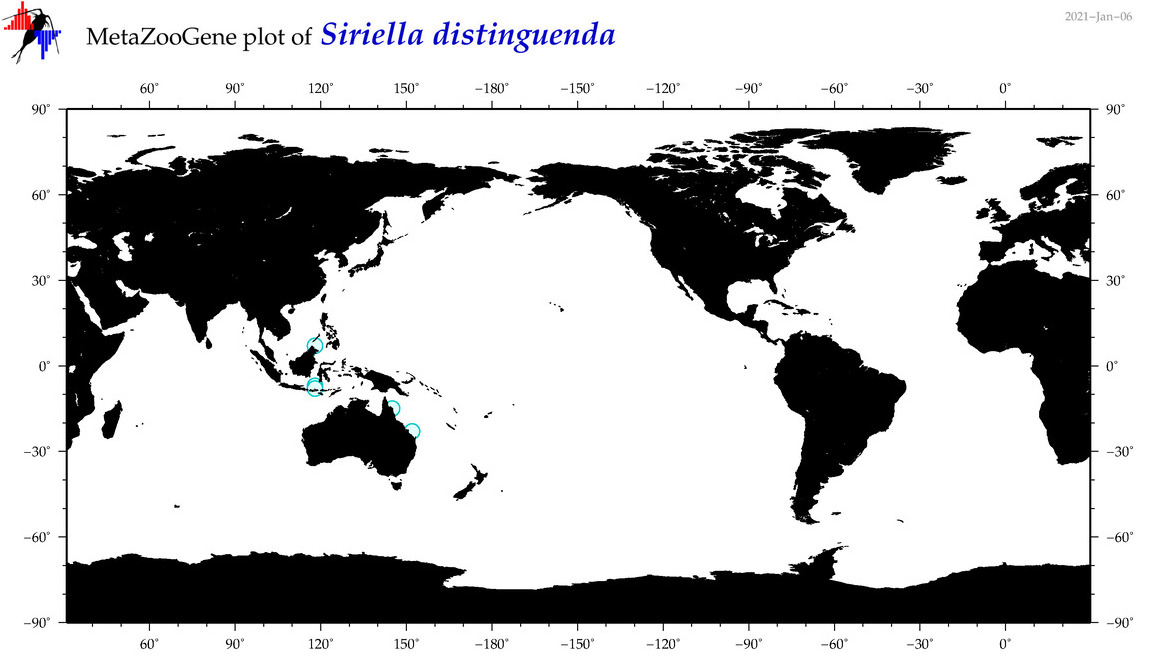 |
accepted
T4068209
R:1:0:0:0 |
| Siriella gracilis |
Species
(77) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
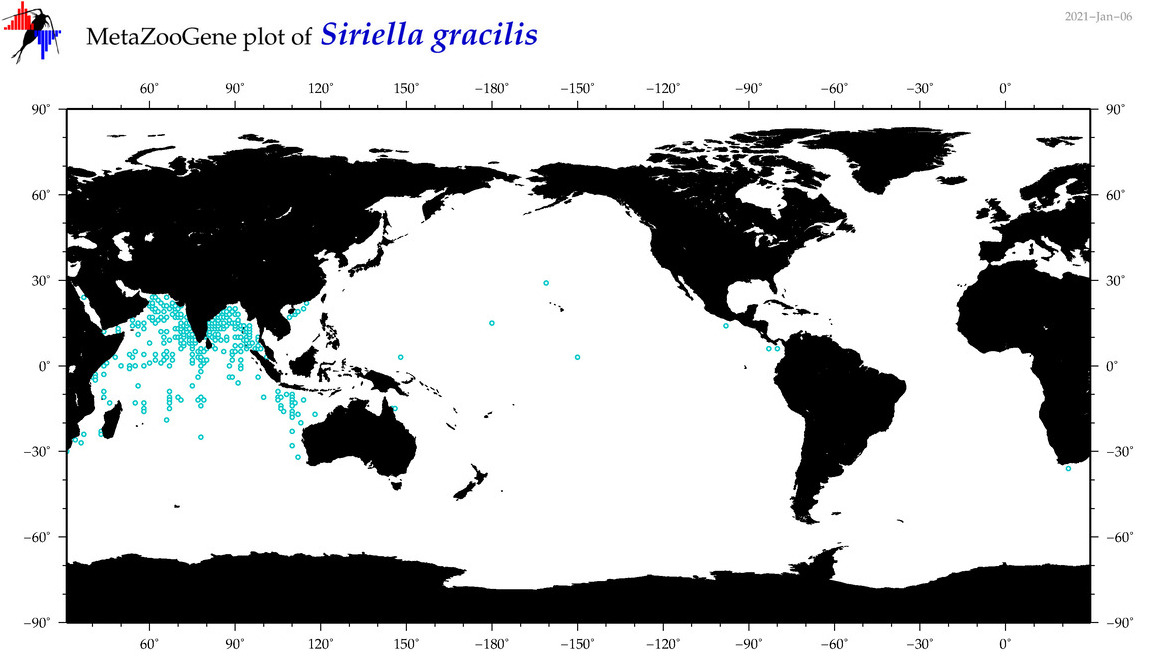 |
accepted
T4007205
R:1:0:0:0 |
| Siriella hanseni |
Species
(78) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
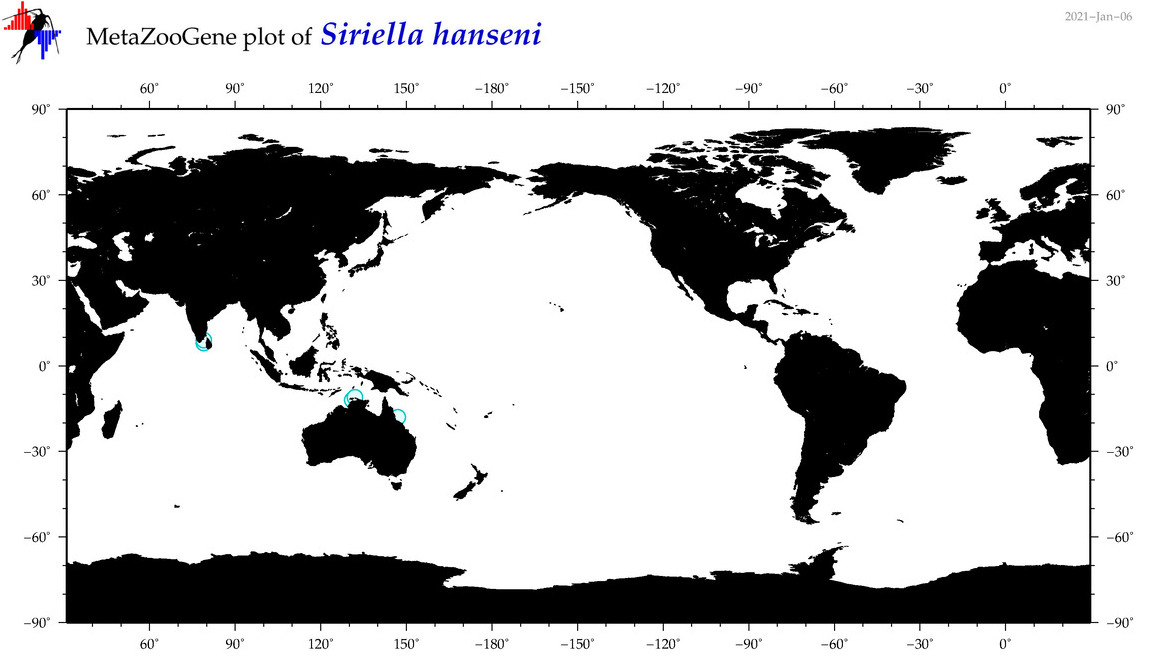 |
accepted
T4068212
R:1:0:0:0 |
| Siriella inornata |
Species
(79) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
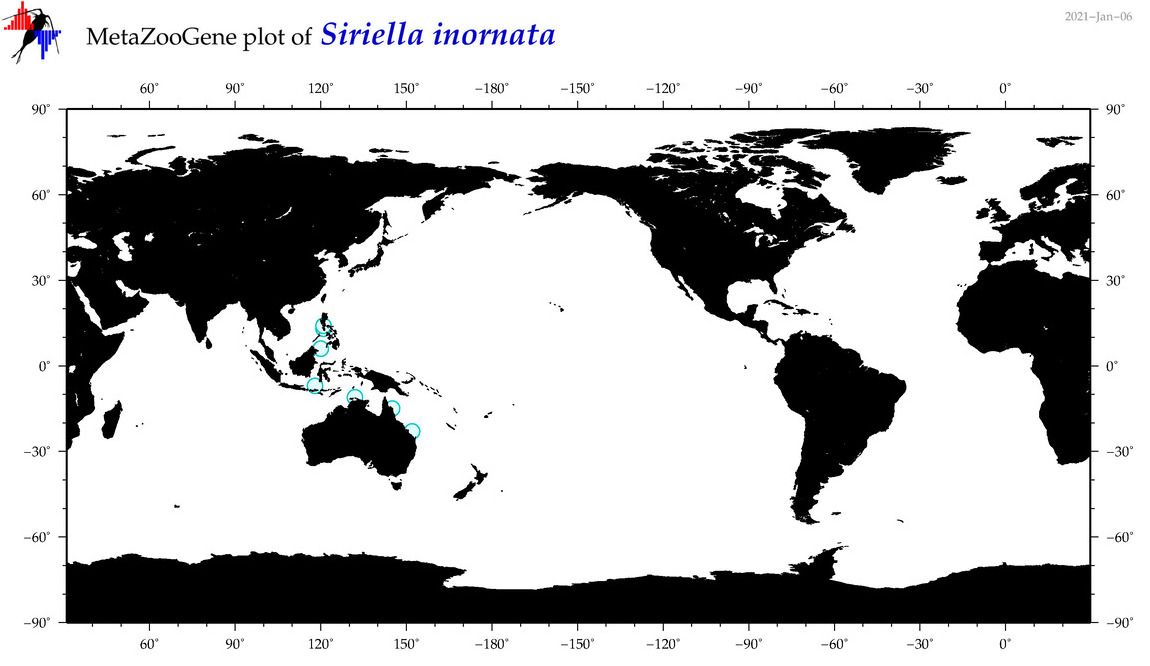 |
accepted
T4068213
R:1:0:0:0 |
| Siriella lacertilis |
Species
(80) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4068216
R:1:0:0:0 |
| Siriella media |
Species
(81) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
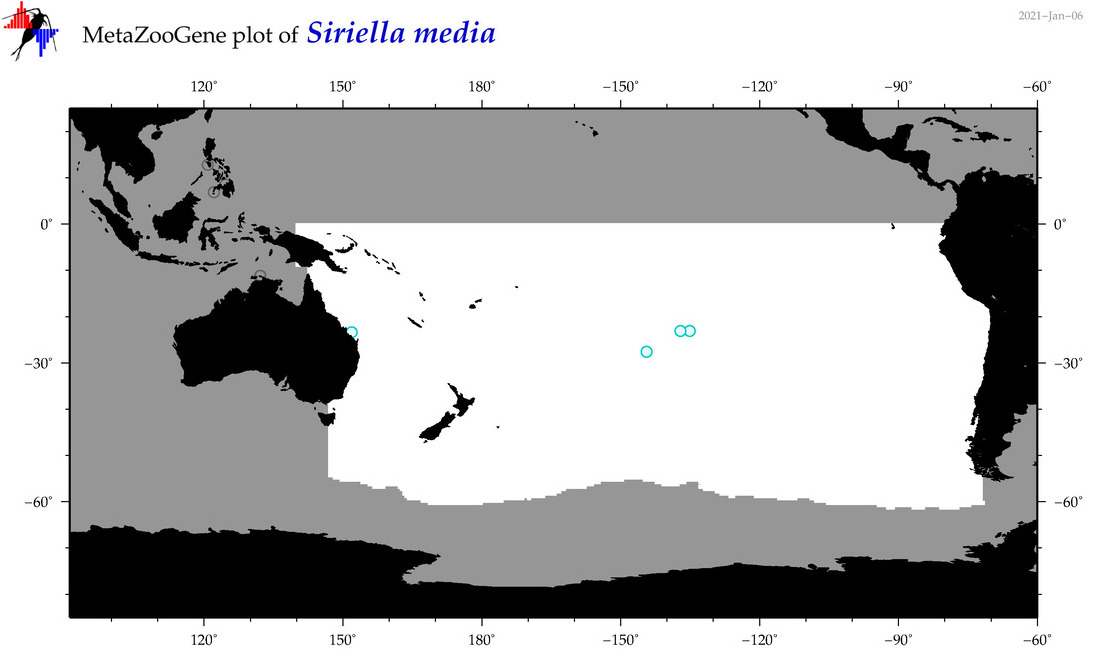 |
accepted
T4068219
R:1:0:0:0 |
| Siriella nodosa |
Species
(82) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
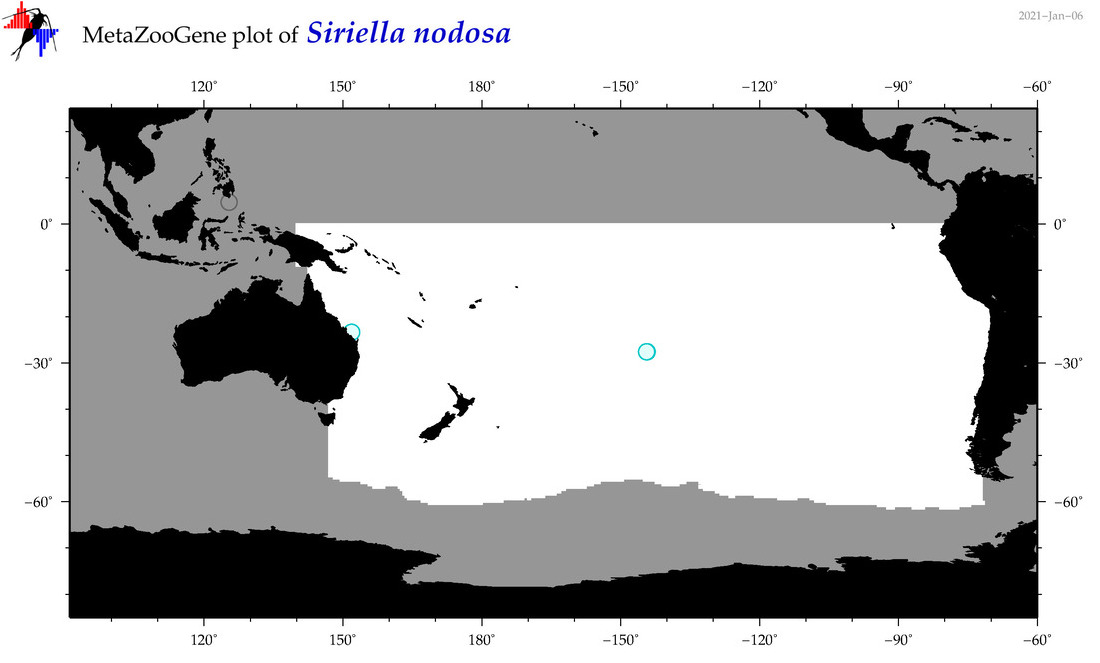 |
accepted
T4068220
R:1:0:0:0 |
| Siriella quadrispinosa |
Species
(83) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
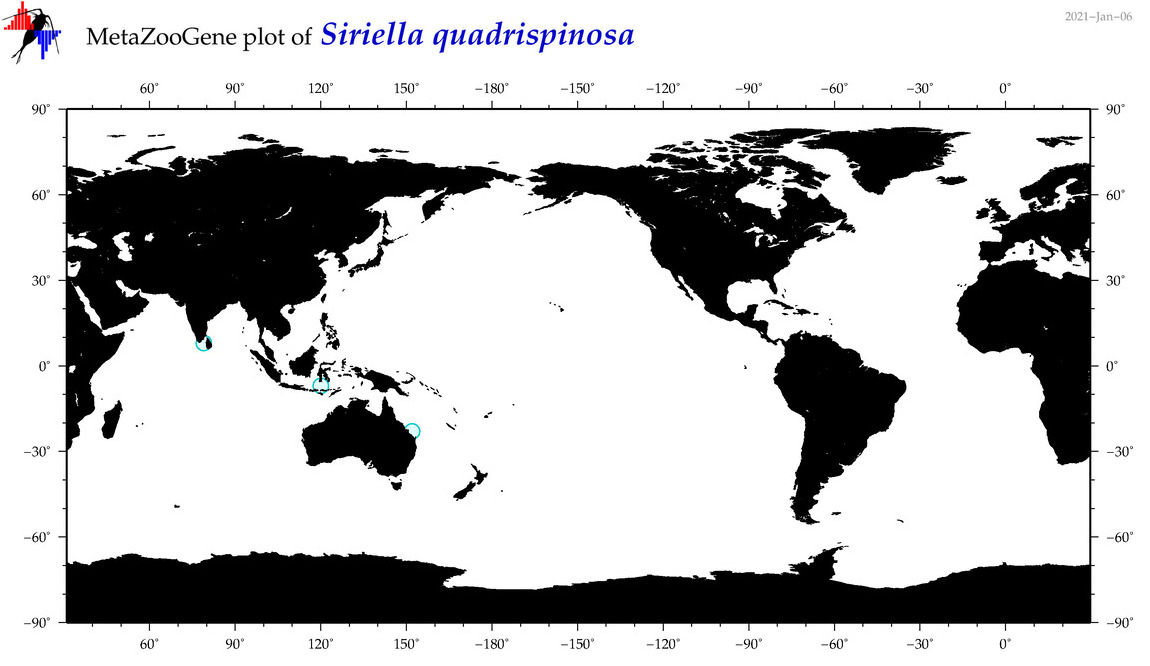 |
accepted
T4068226
R:1:0:0:0 |
| Siriella thompsonii |
Species
(84) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
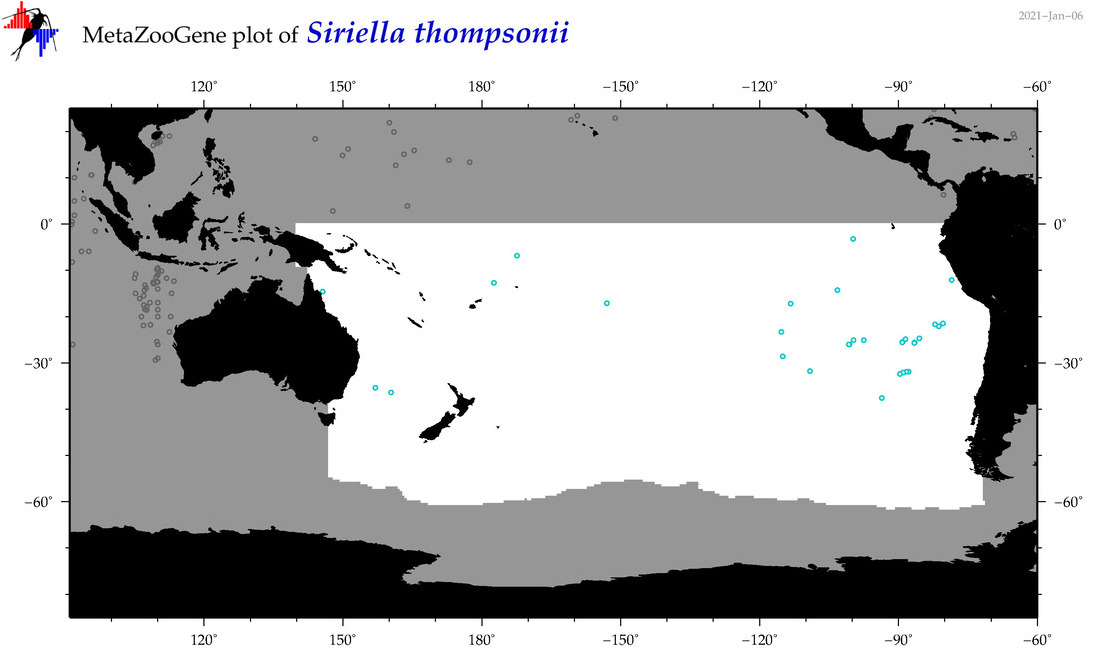 |
accepted
T4007208
R:1:0:0:0 |
| Siriella vincenti |
Species
(85) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4007209
R:1:0:0:0 |
| Siriella vulgaris |
Species
(86) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
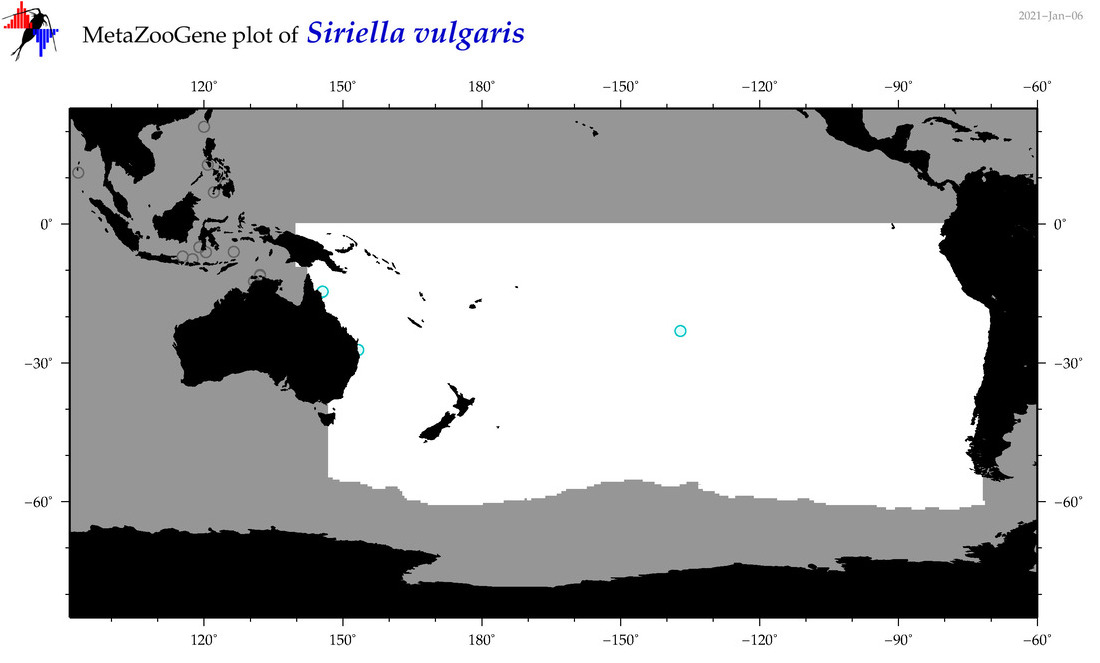 |
accepted
T4068231
R:1:0:0:0 |
| Tenagomysis australis |
Species
(87) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
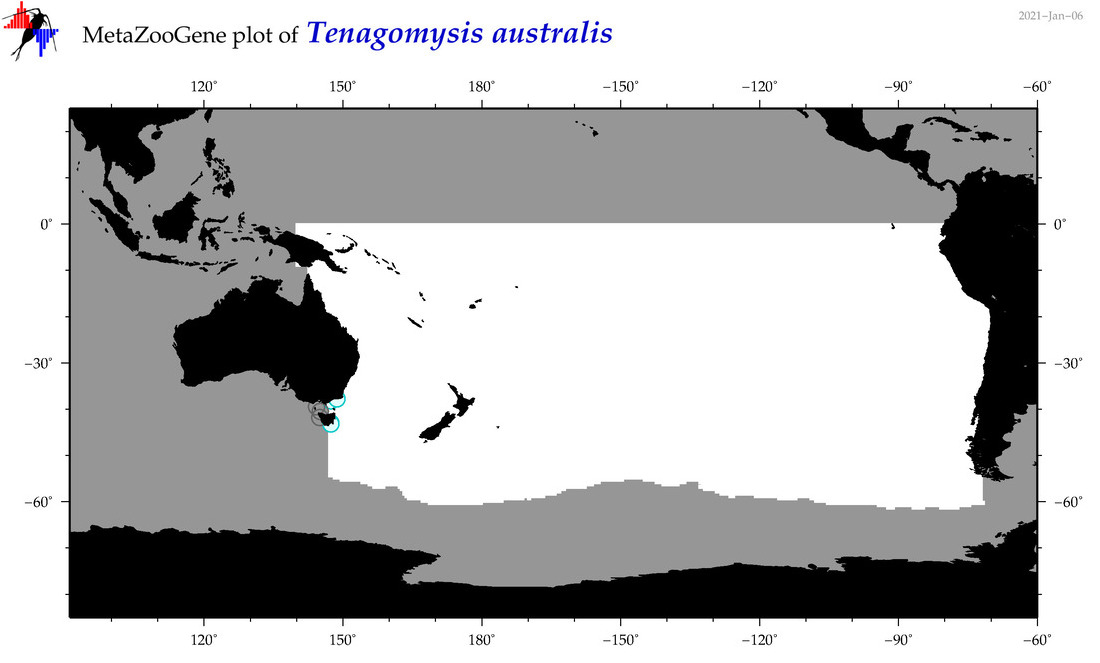 |
accepted
T4015287
R:1:0:0:0 |
| Tenagomysis bruniensis |
Species
(88) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4070001
R:1:0:0:0 |
| Tenagomysis macropsis |
Species
(89) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
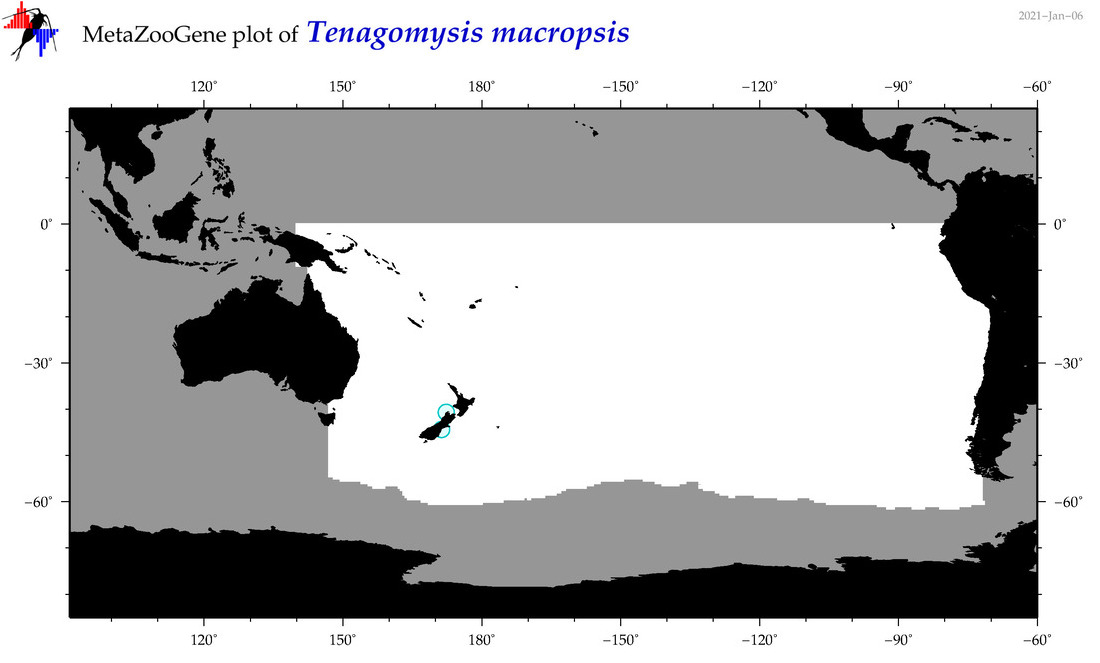 |
accepted
T4070003
R:1:0:0:0 |
| Tenagomysis robusta |
Species
(90) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
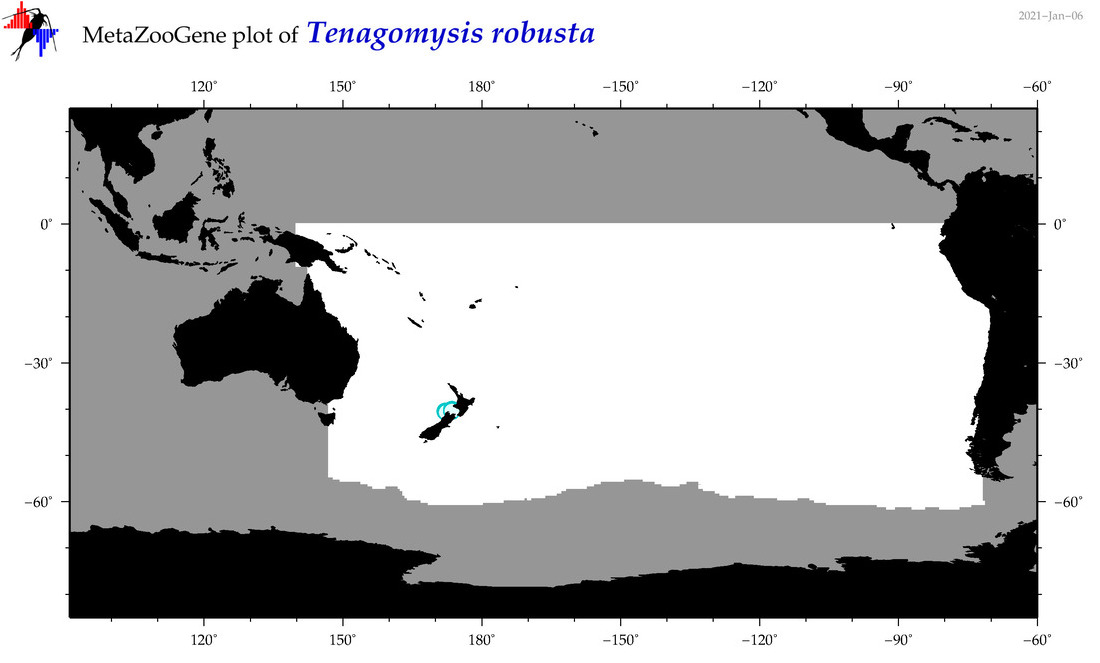 |
accepted
T4070005
R:1:0:0:0 |
| Tenagomysis scotti |
Species
(91) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
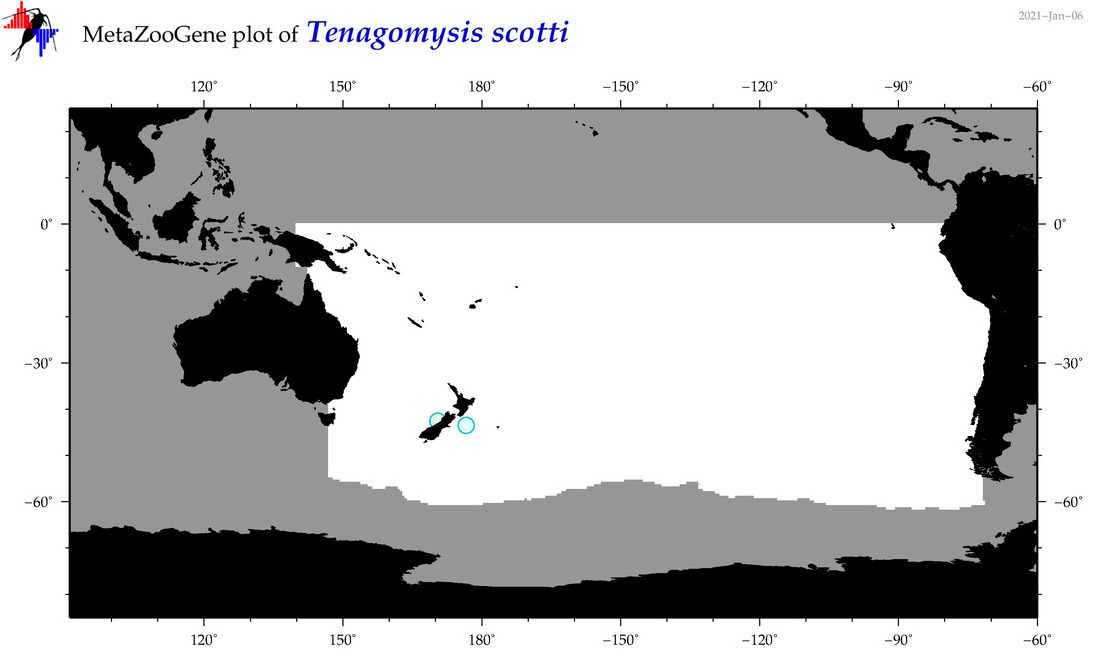 |
accepted
T4070006
R:1:0:0:0 |
| Tenagomysis similis |
Species
(92) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4099981
R:1:0:0:0 |
| Tenagomysis tasmaniae |
Species
(93) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
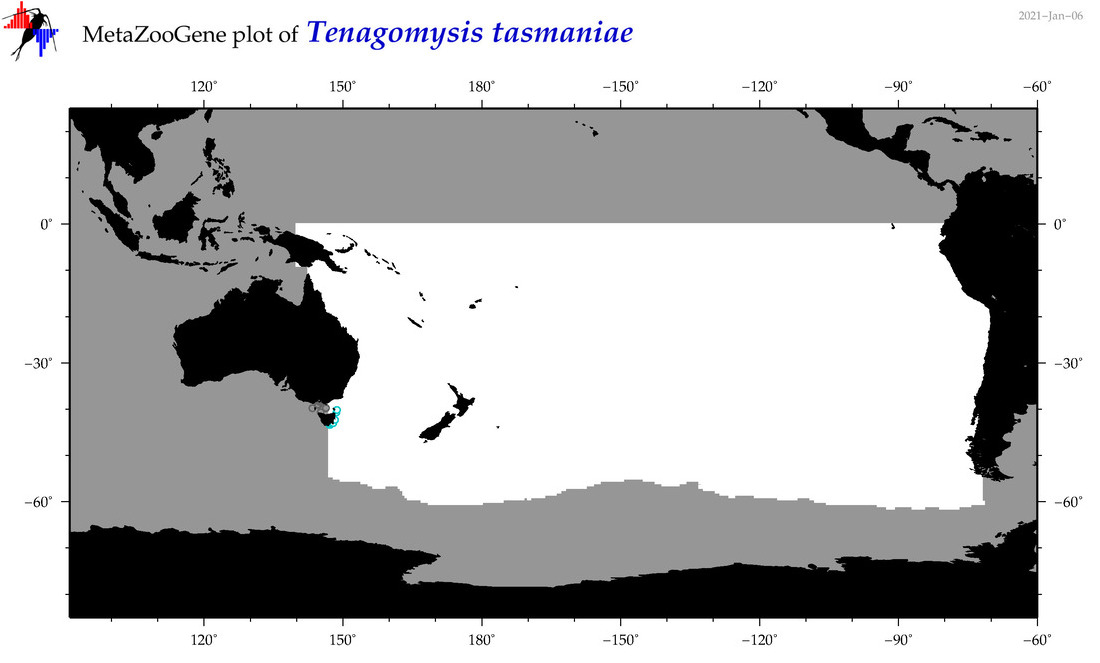 |
accepted
T4007210
R:1:0:0:0 |
| Tenagomysis tenuipes |
Species
(94) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
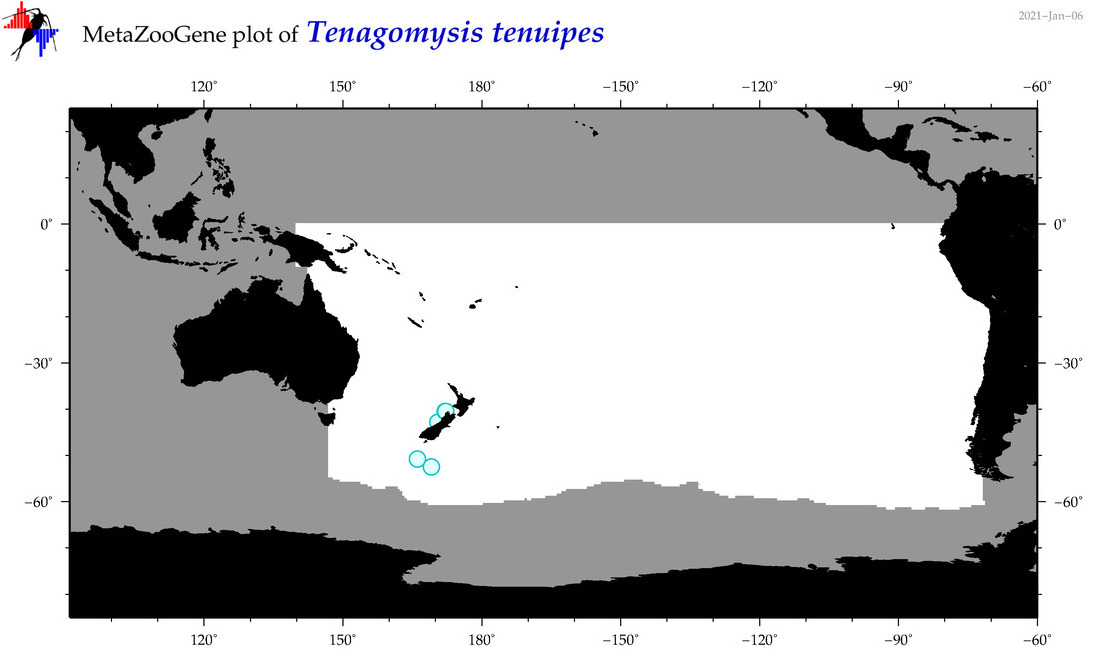 |
accepted
T4070007
R:1:0:0:0 |
| Tenagomysis thomsoni |
Species
(95) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
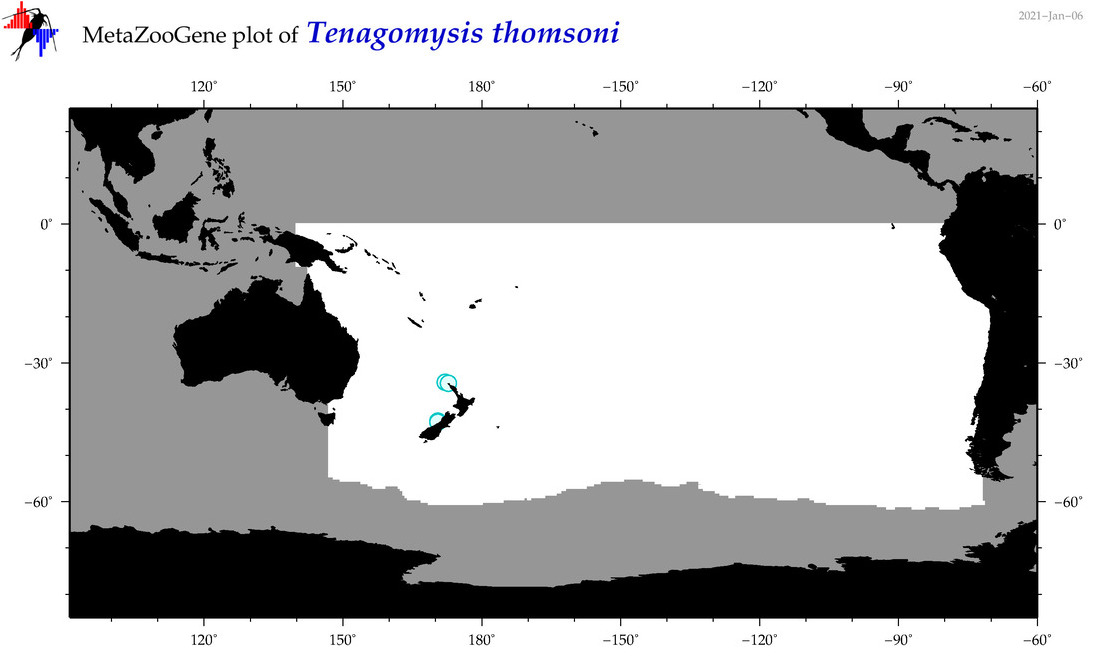 |
accepted
T4070008
R:1:0:0:0 |