Taxa Name
| Taxa
Level | Barcode
Data? | Number of
Barcodes
from
any ocean | Number of
Barcodes
from this region | Barcode Locations and
Species Occurence Map | MZGdb Atlas
v2026-m01-01
2026-Jan-24 |
| Alamprops comatus |
Species
(1) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
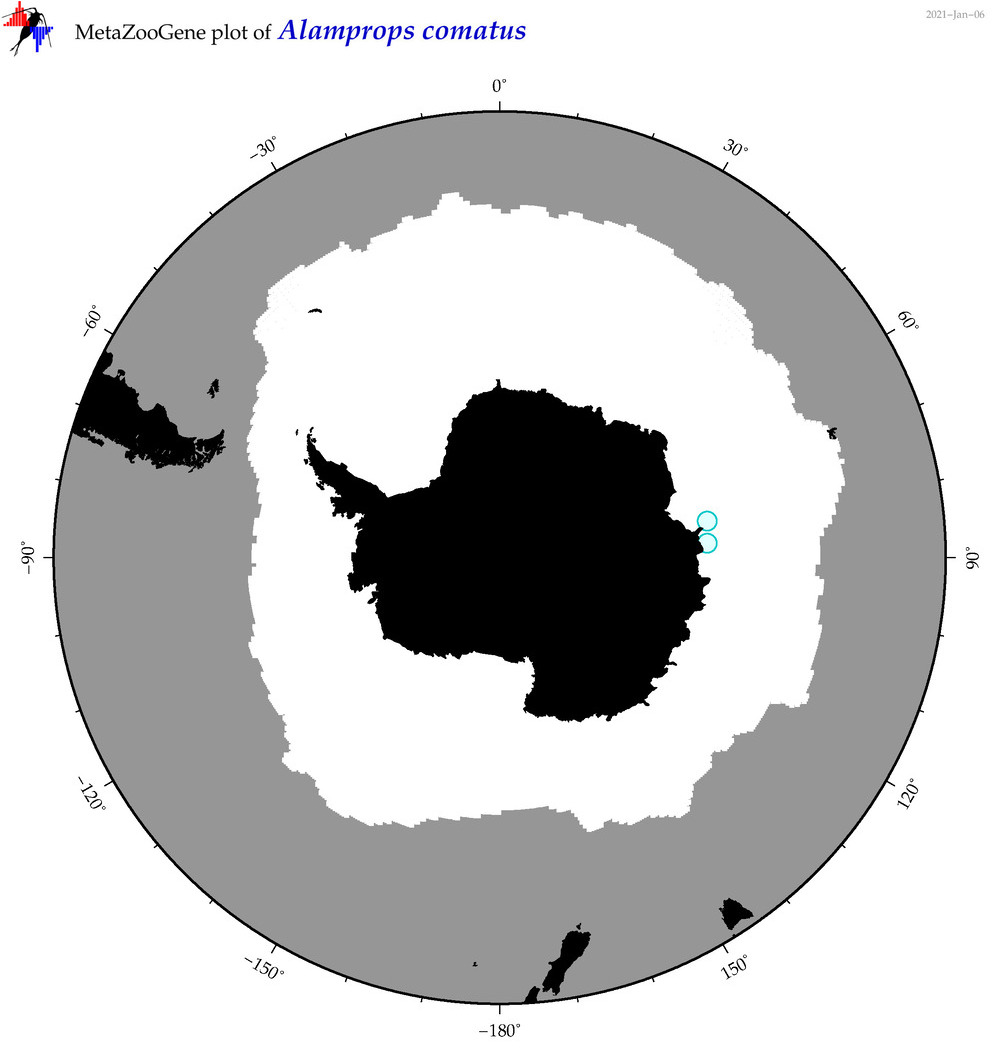 |
accepted
T4042444
R:1:0:0:0 |
| Atlantocuma (Atlantocuma) bidentatum |
Species
(2) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4094801
R:1:0:0:0 |
| Bathycuma capense |
Species
(3) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
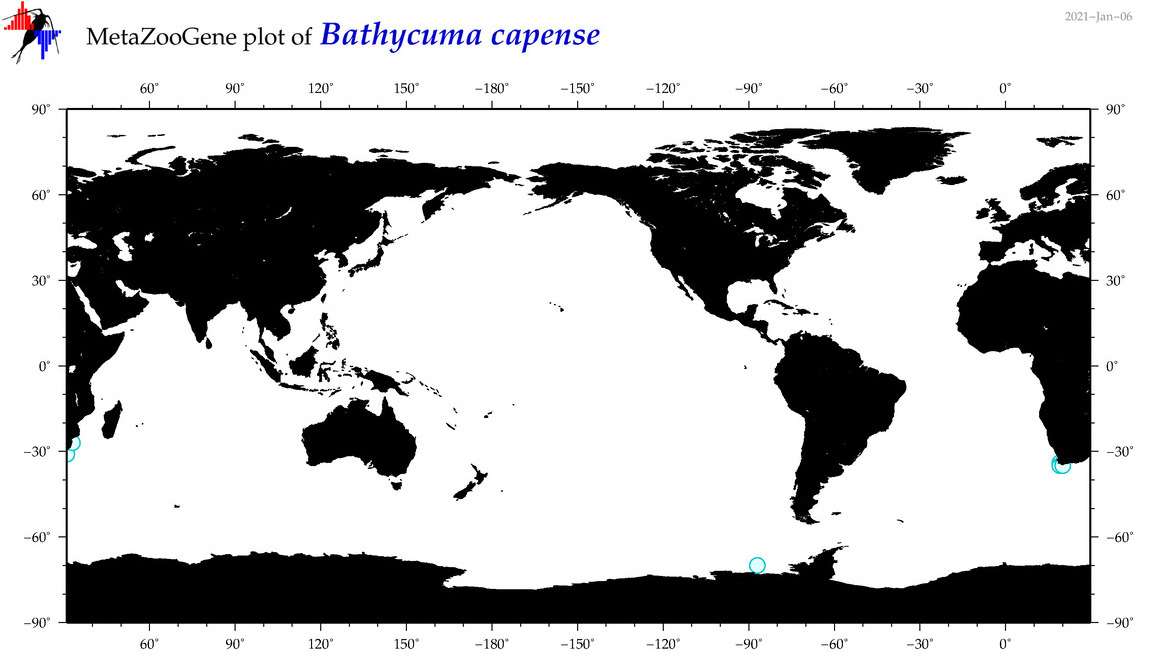 |
accepted
T4045195
R:1:0:0:0 |
| Campylaspis alisae |
Species
(4) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4046526
R:1:0:0:0 |
| Campylaspis antarctica |
Species
(5) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
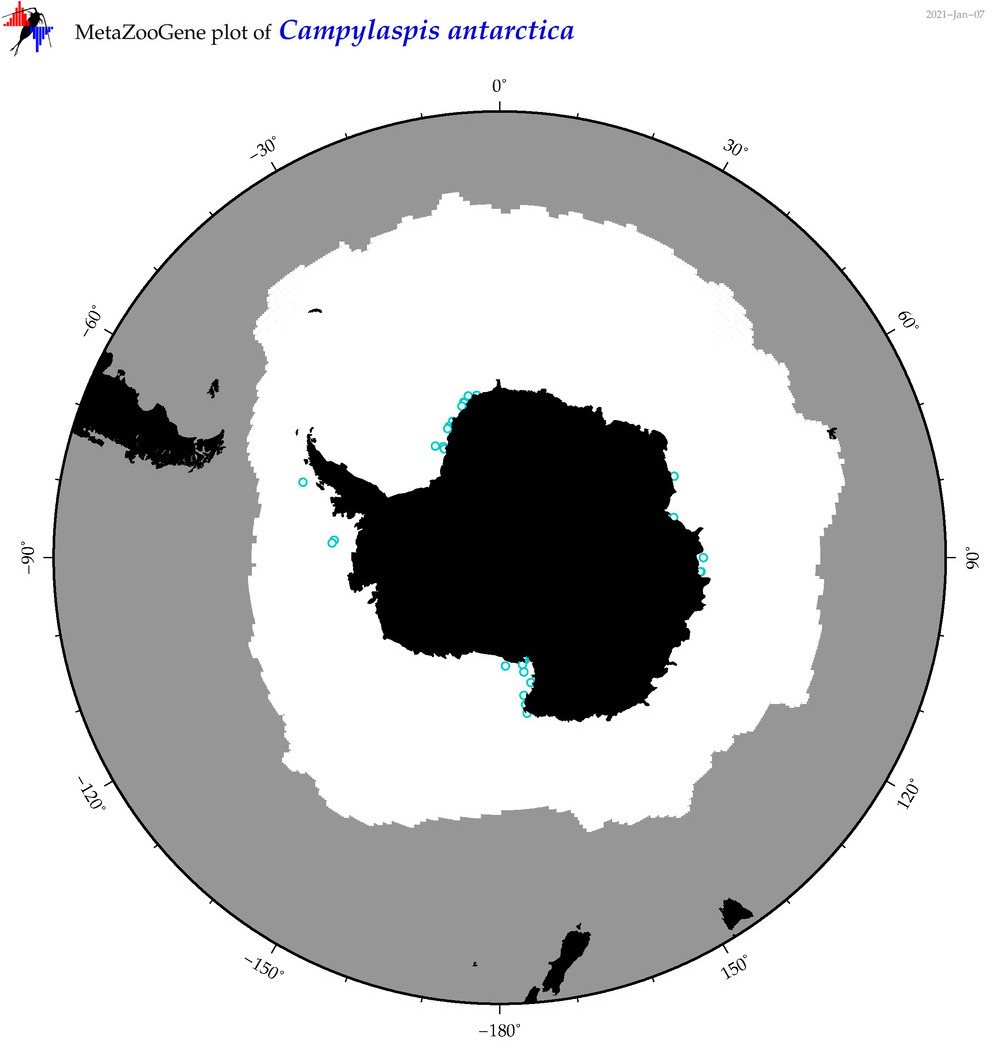 |
accepted
T4046529
R:1:0:0:0 |
| Campylaspis breviramis |
Species
(6) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
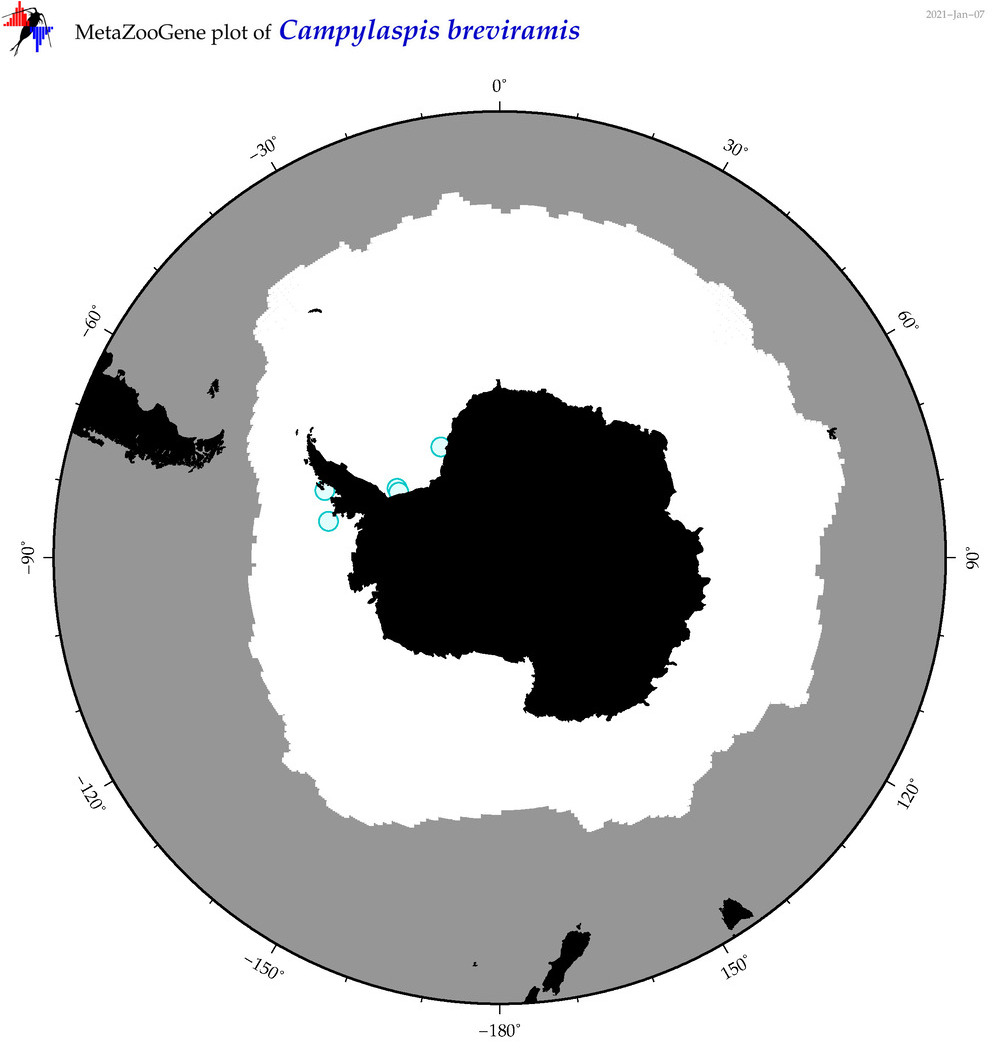 |
accepted
T4046539
R:1:0:0:0 |
| Campylaspis excavata |
Species
(7) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
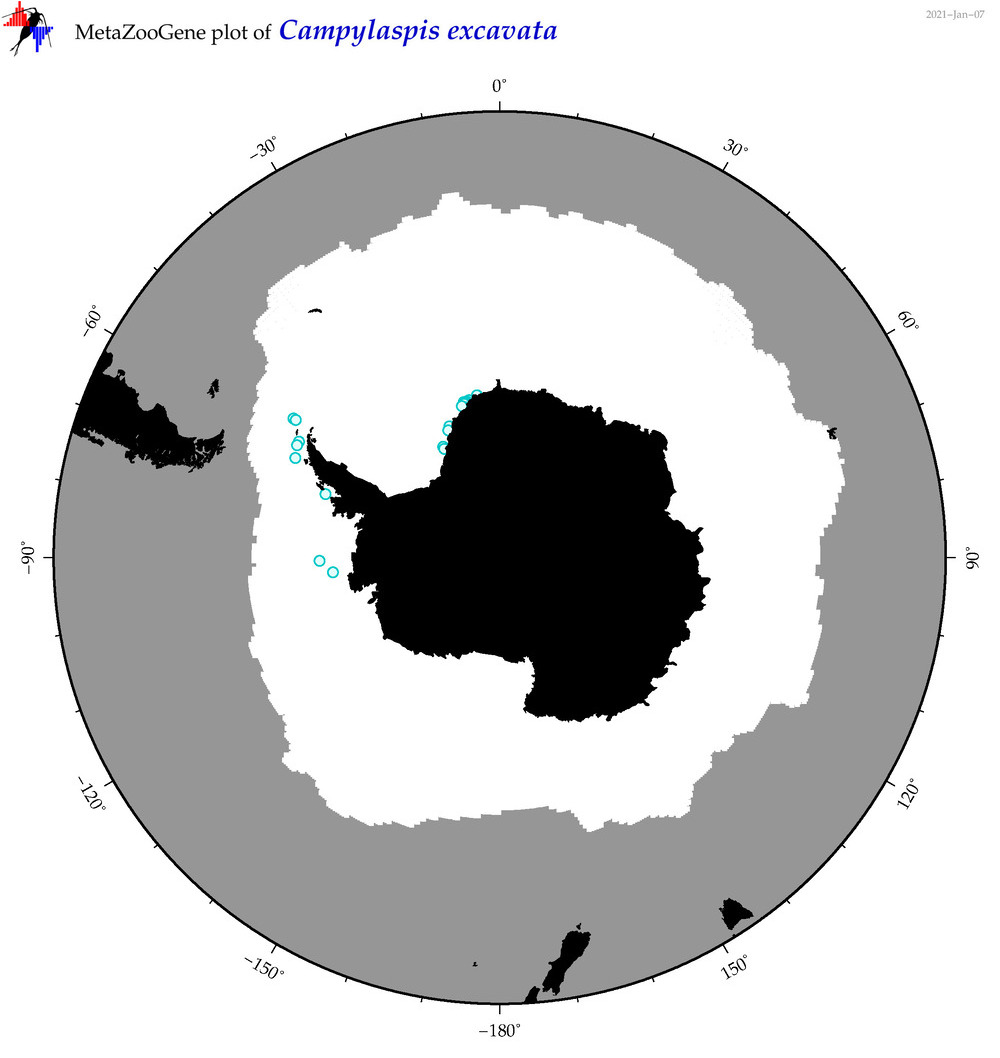 |
accepted
T4046550
R:1:0:0:0 |
| Campylaspis frigida |
Species
(8) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
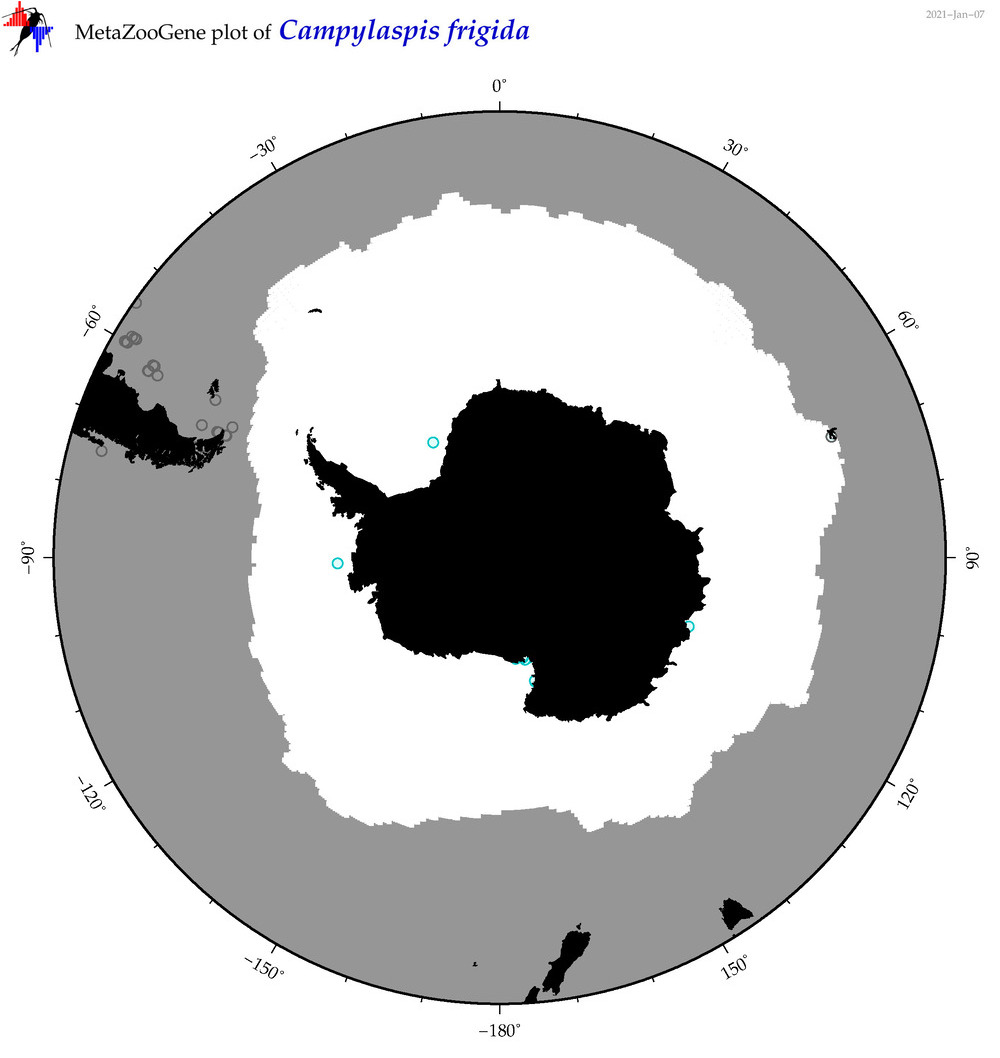 |
accepted
T4046551
R:1:0:0:0 |
| Campylaspis heterotuberculata |
Species
(9) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
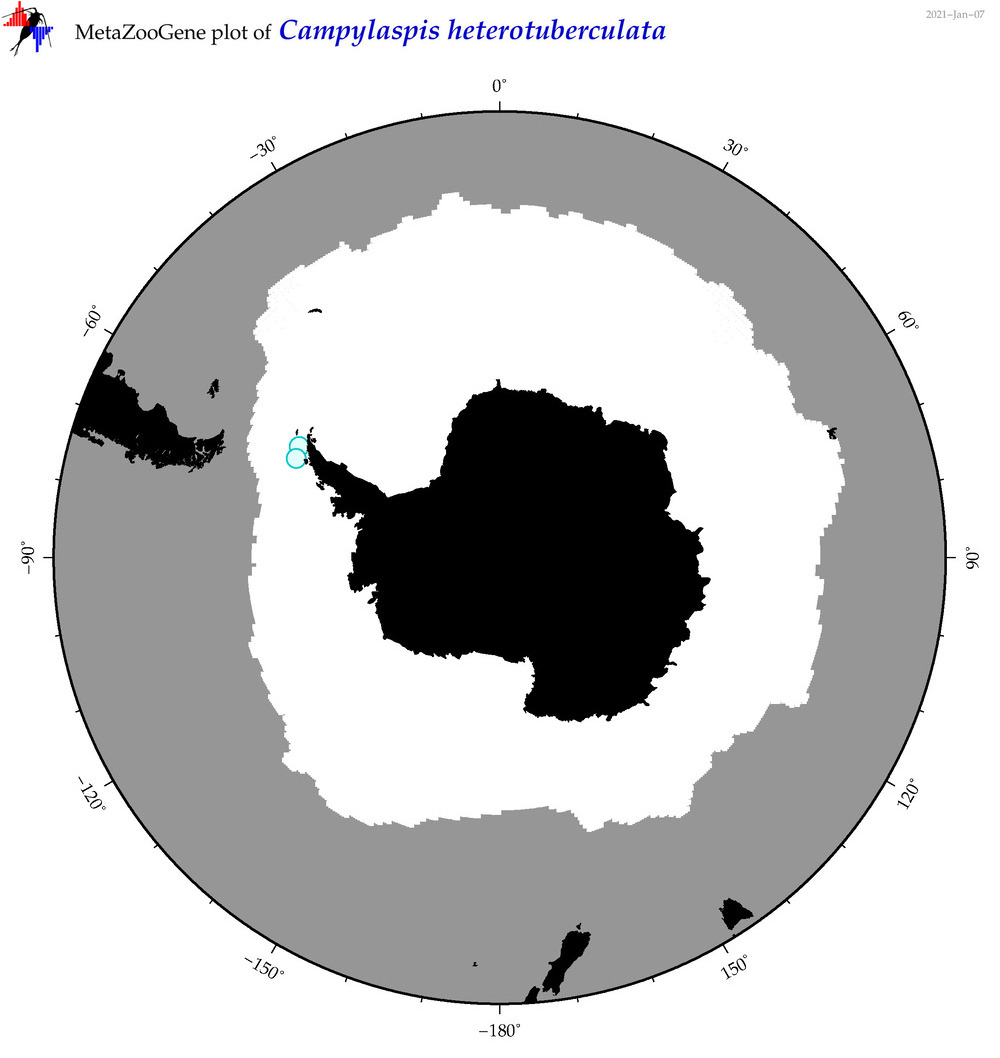 |
accepted
T4046563
R:1:0:0:0 |
| Campylaspis johnstoni |
Species
(10) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
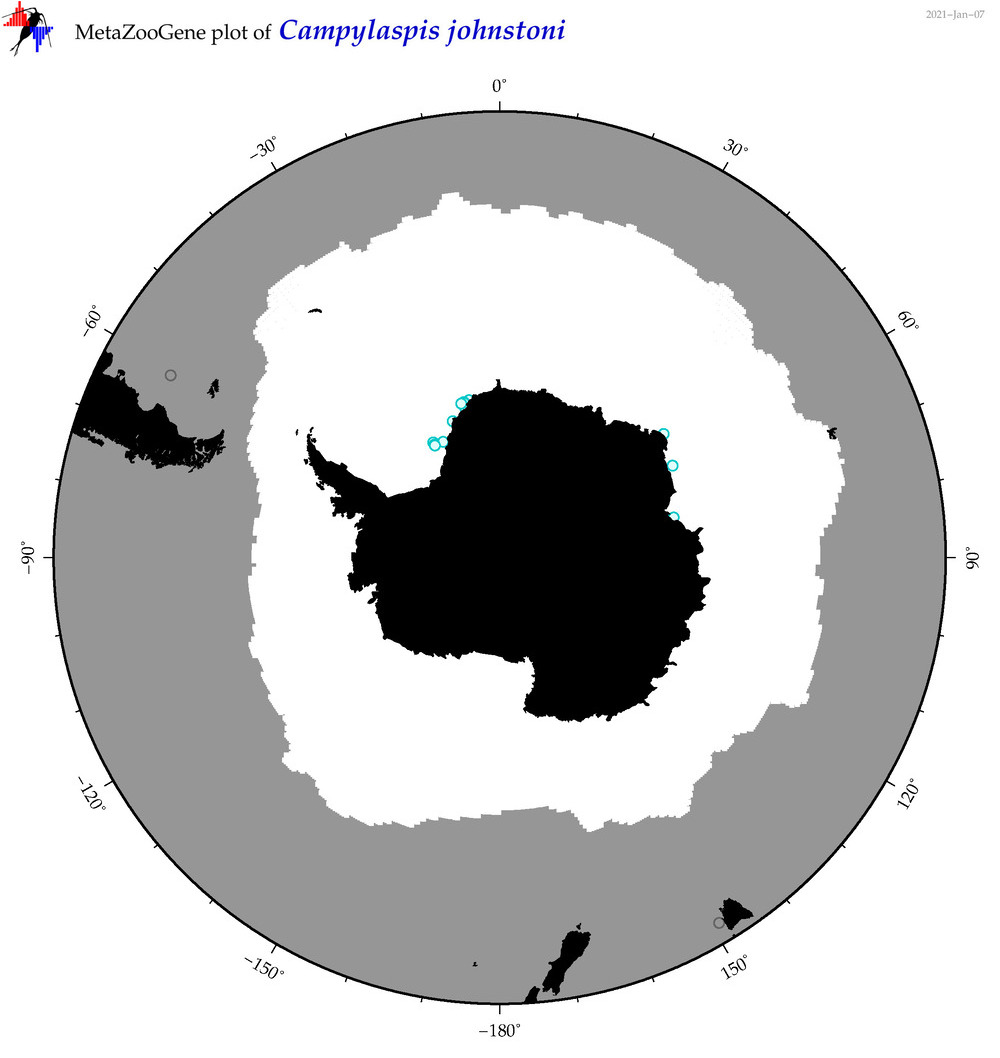 |
accepted
T4046569
R:1:0:0:0 |
| Campylaspis ledoyeri |
Species
(11) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
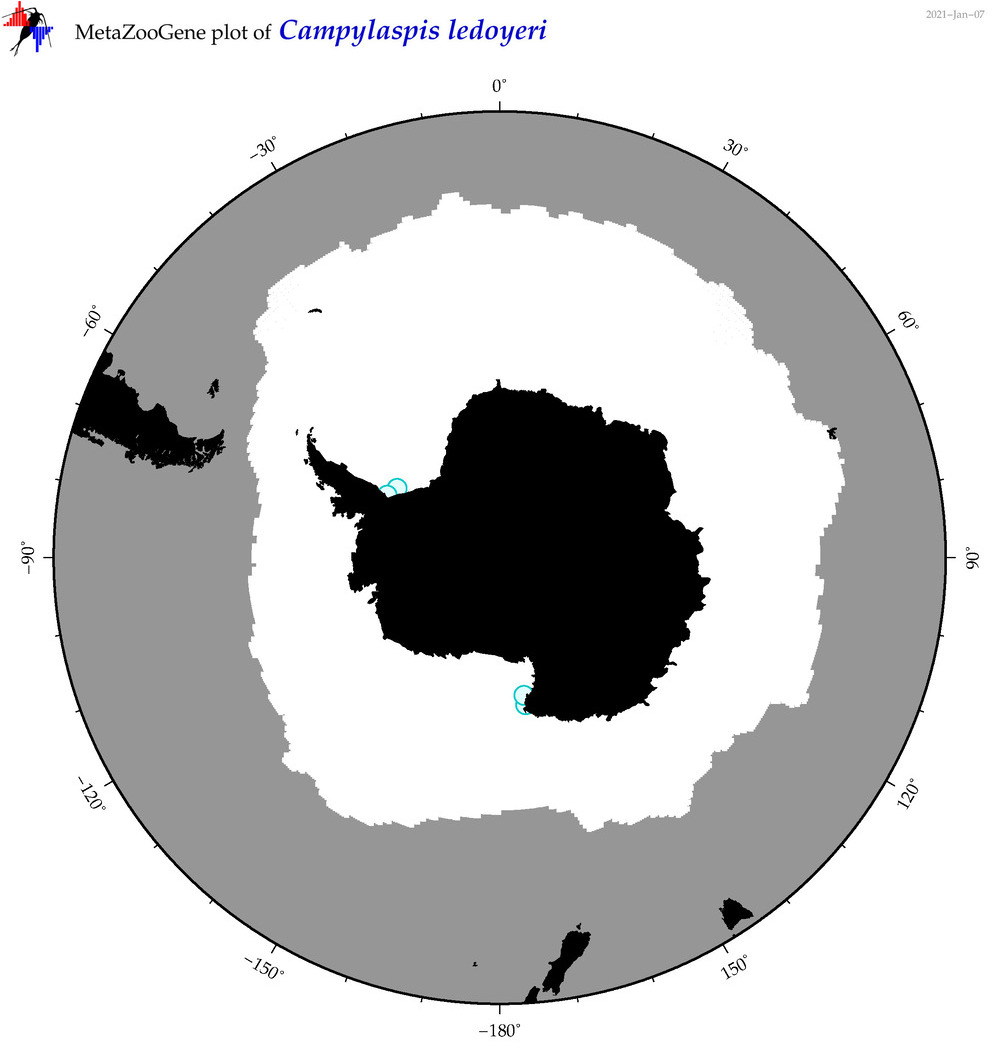 |
accepted
T4046576
R:1:0:0:0 |
| Campylaspis maculata |
Species
(12) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
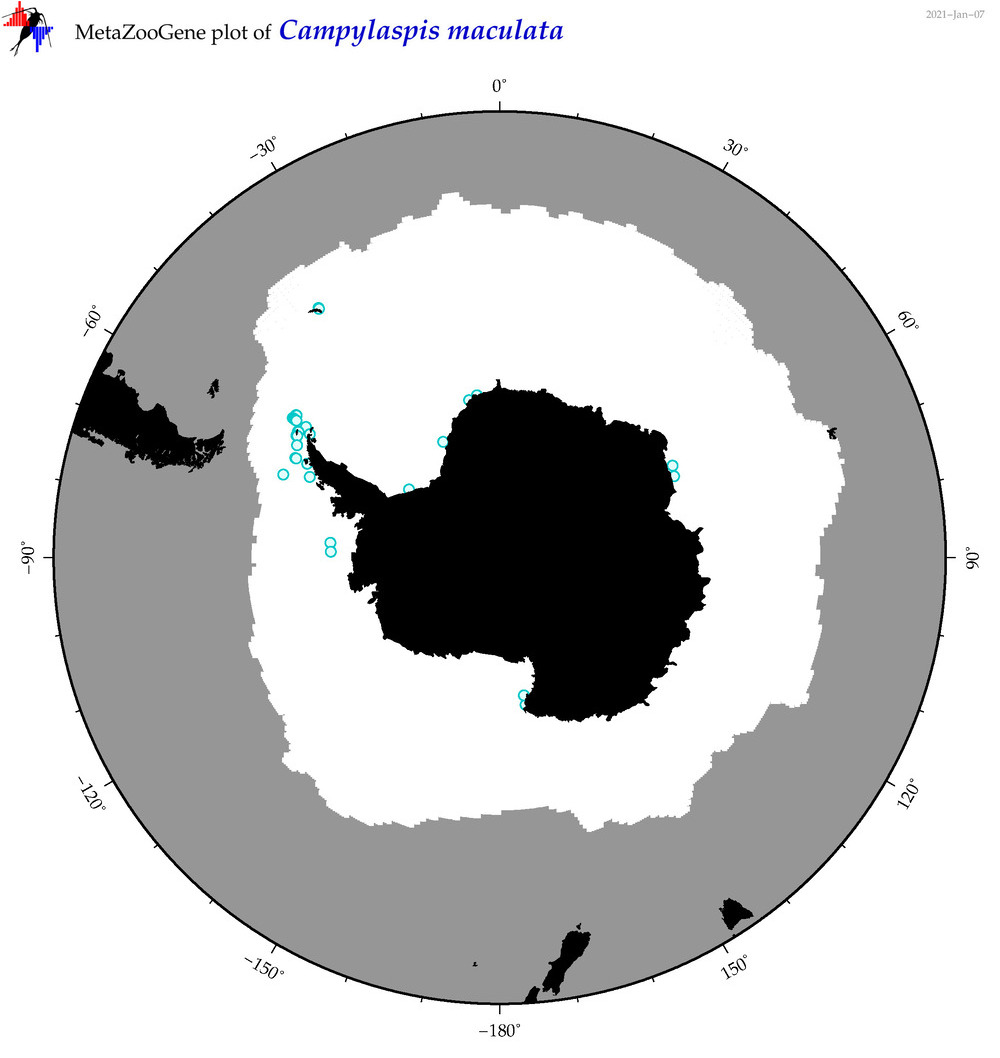 |
accepted
T4046582
R:1:0:0:0 |
| Campylaspis nodulosa |
Species
(13) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
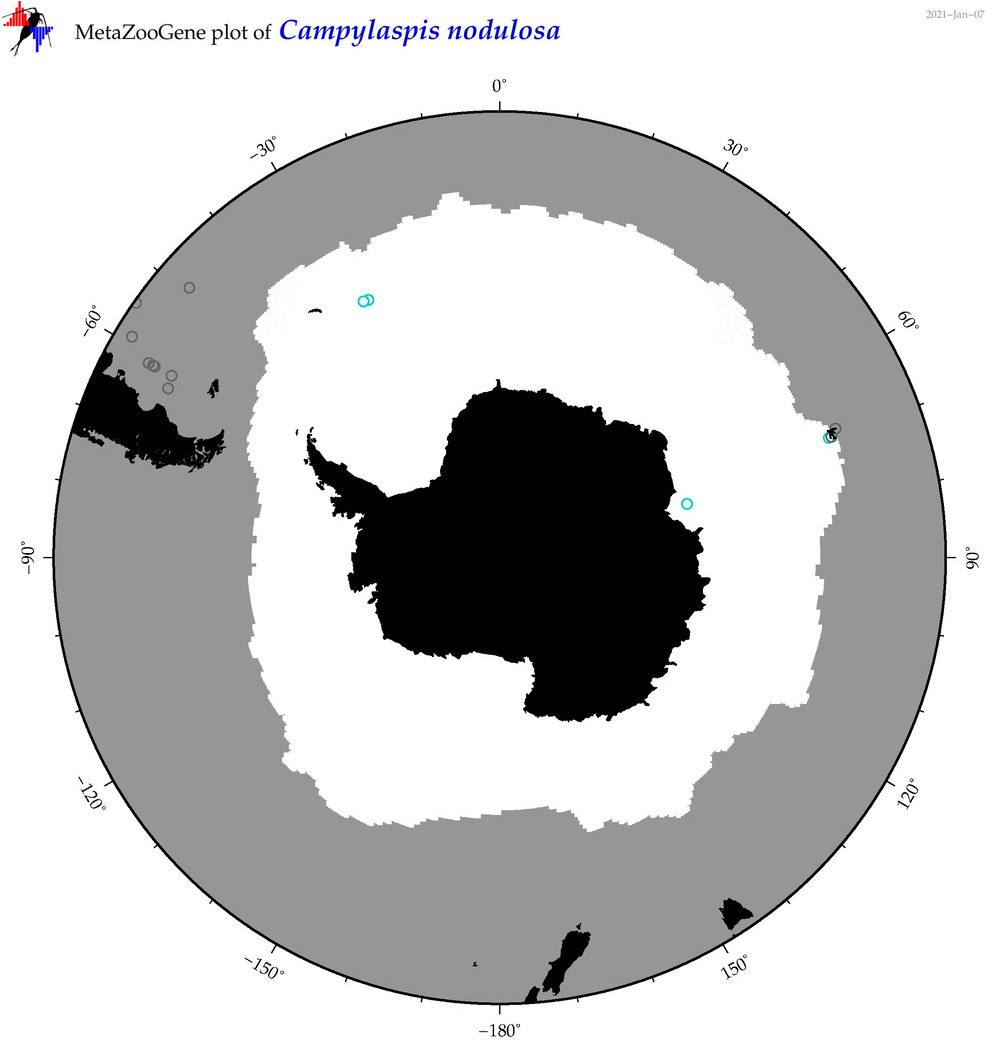 |
accepted
T4046593
R:1:0:0:0 |
| Campylaspis quadridentata |
Species
(14) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
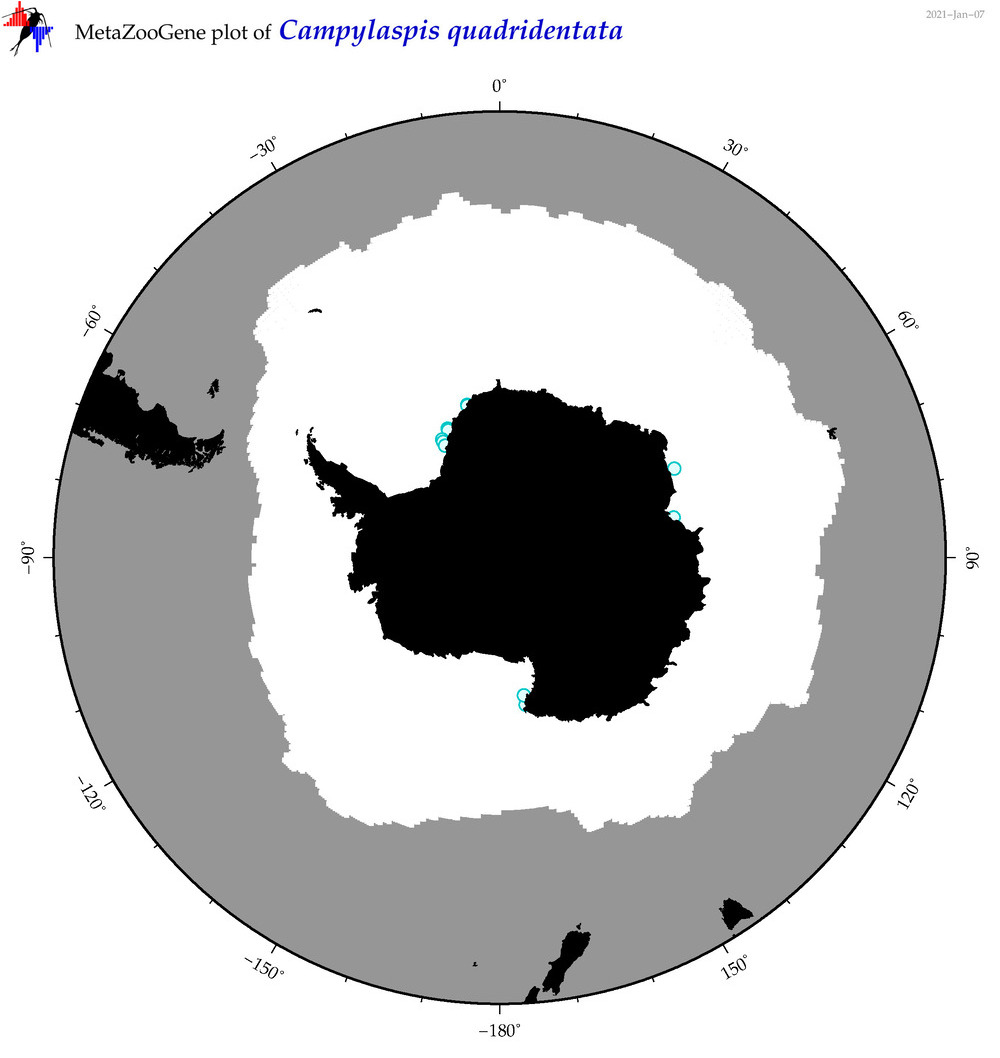 |
accepted
T4046607
R:1:0:0:0 |
| Campylaspis quadriplicata |
Species
(15) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
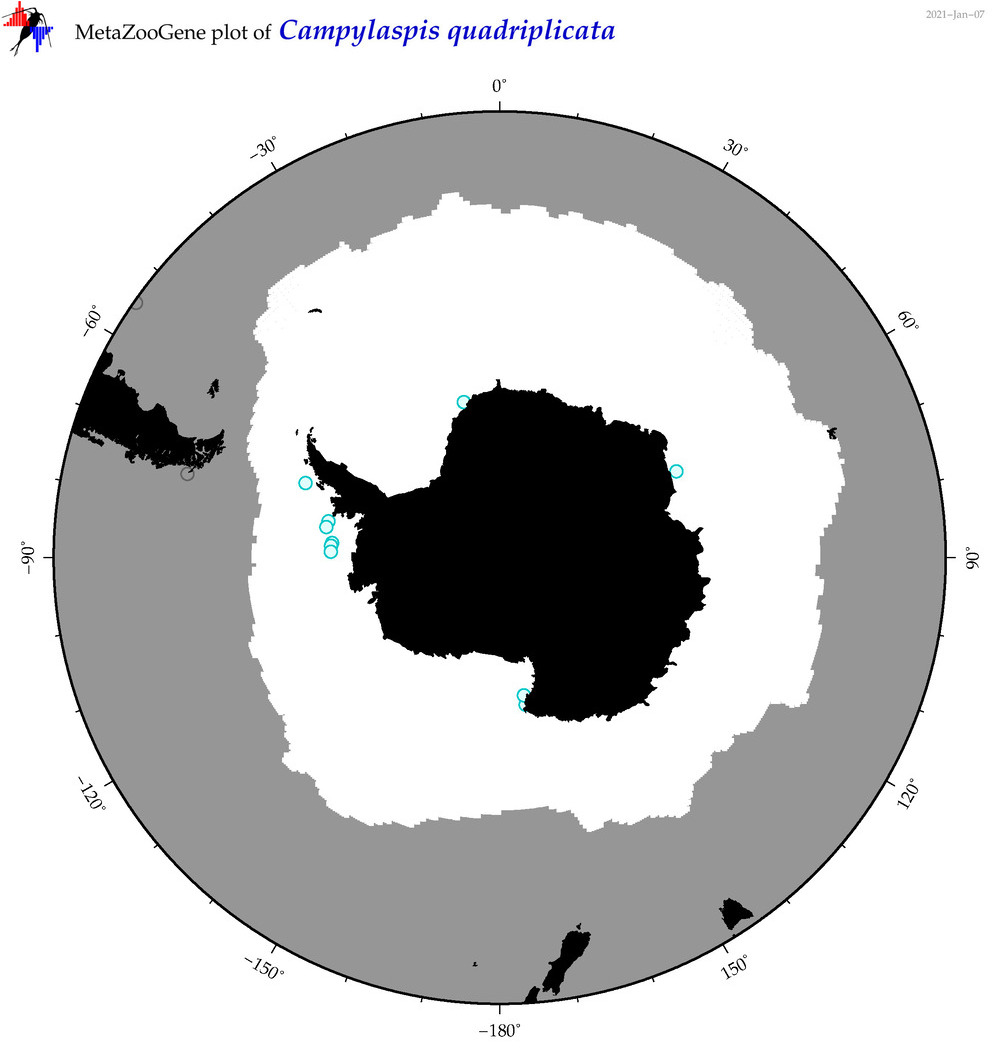 |
accepted
T4046608
R:1:0:0:0 |
| Cumella (Cumella) australis |
Species
(16) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
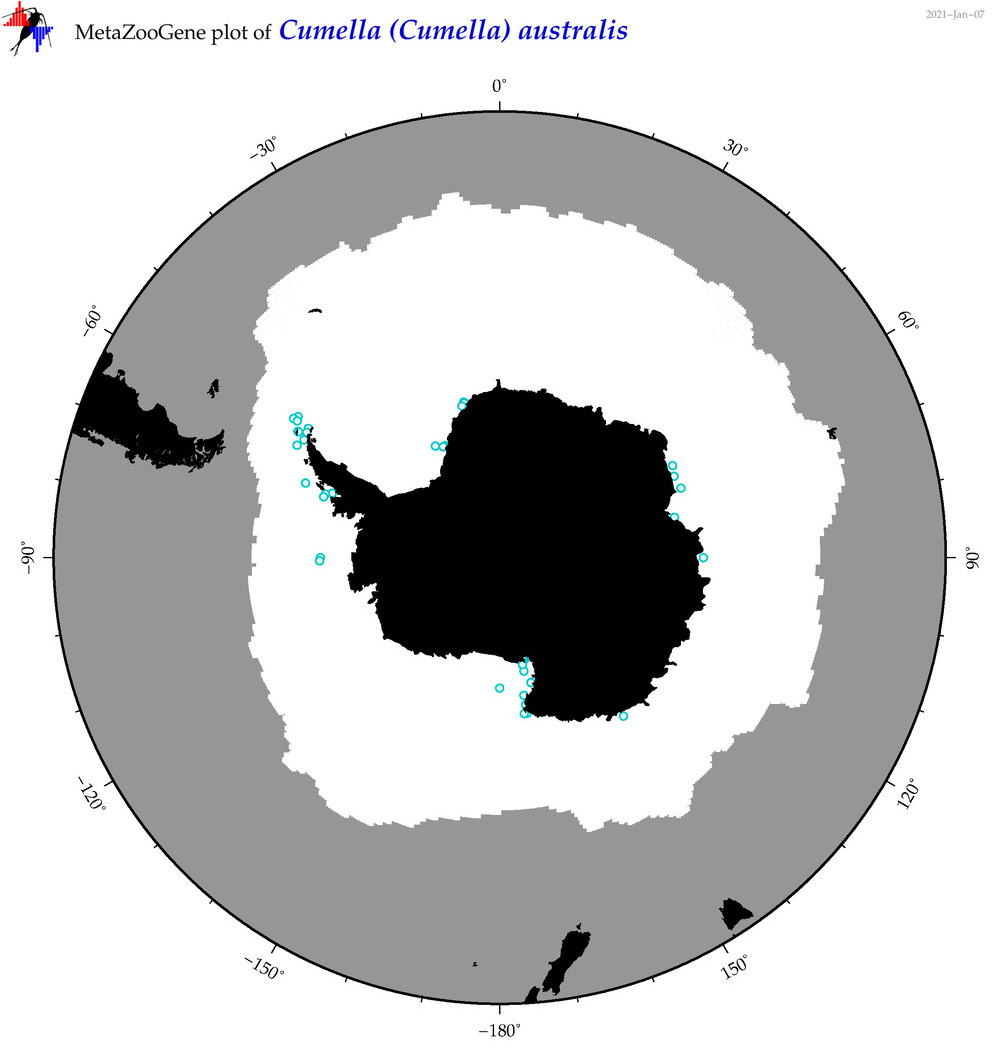 |
accepted
T4094872
R:1:0:0:0 |
| Cumella (Cumella) meridionalis |
Species
(17) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
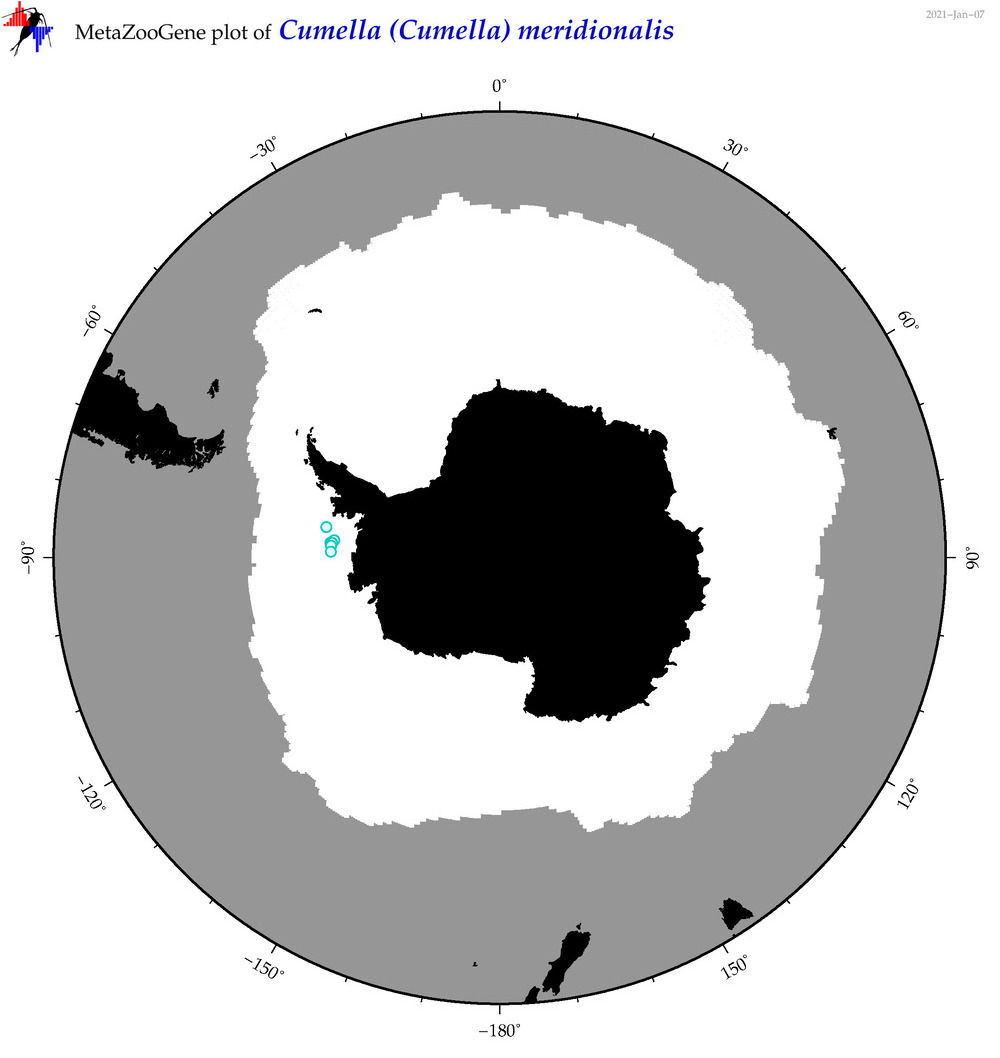 |
accepted
T4094884
R:1:0:0:0 |
| Cumella (Cumella) pectinifera |
Species
(18) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4130745
R:1:0:0:0 |
| Cumella emergens |
Species
(19) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4078024
R:1:0:0:0 |
| Cyclaspis cristulata |
Species
(20) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4048893
R:1:0:0:0 |
| Cyclaspis gigas |
Species
(21) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
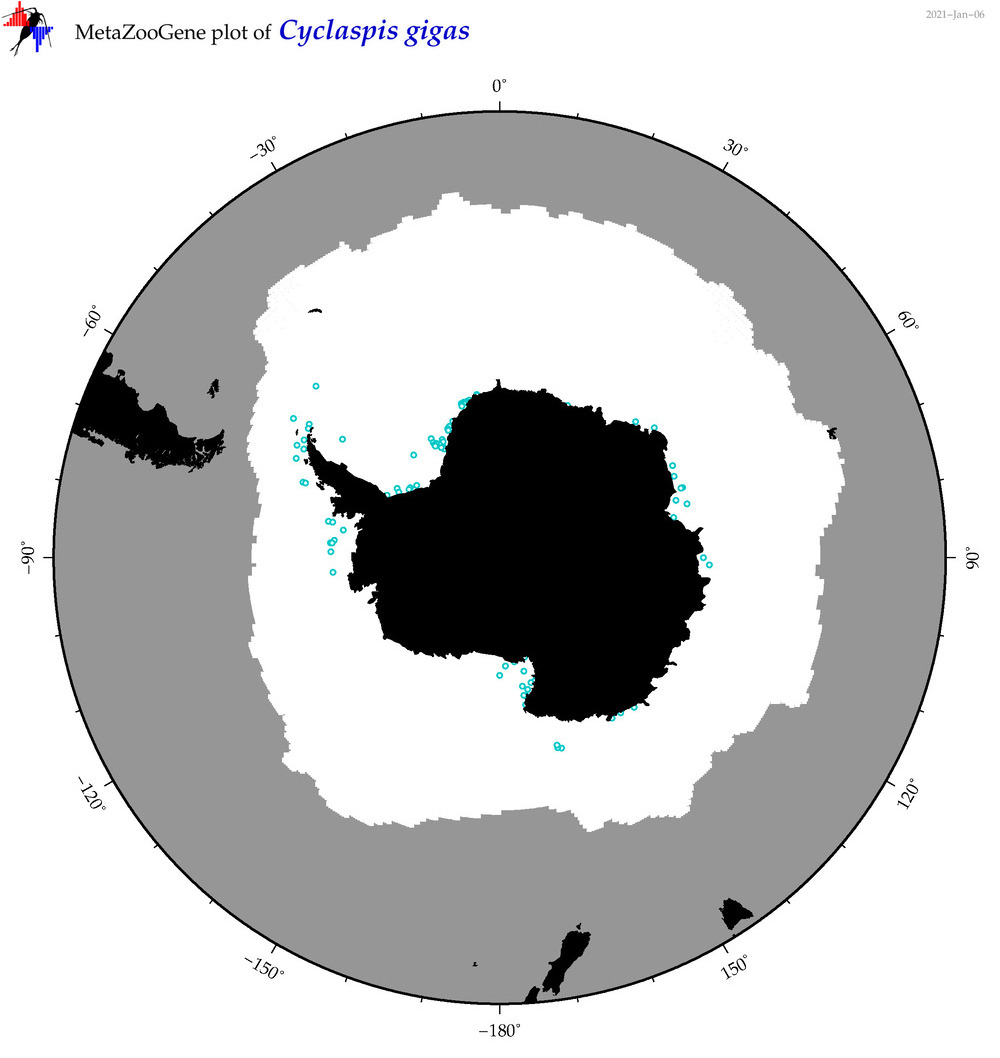 |
accepted
T4048896
R:1:0:0:0 |
| Cyclaspis quadrituberculata |
Species
(22) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
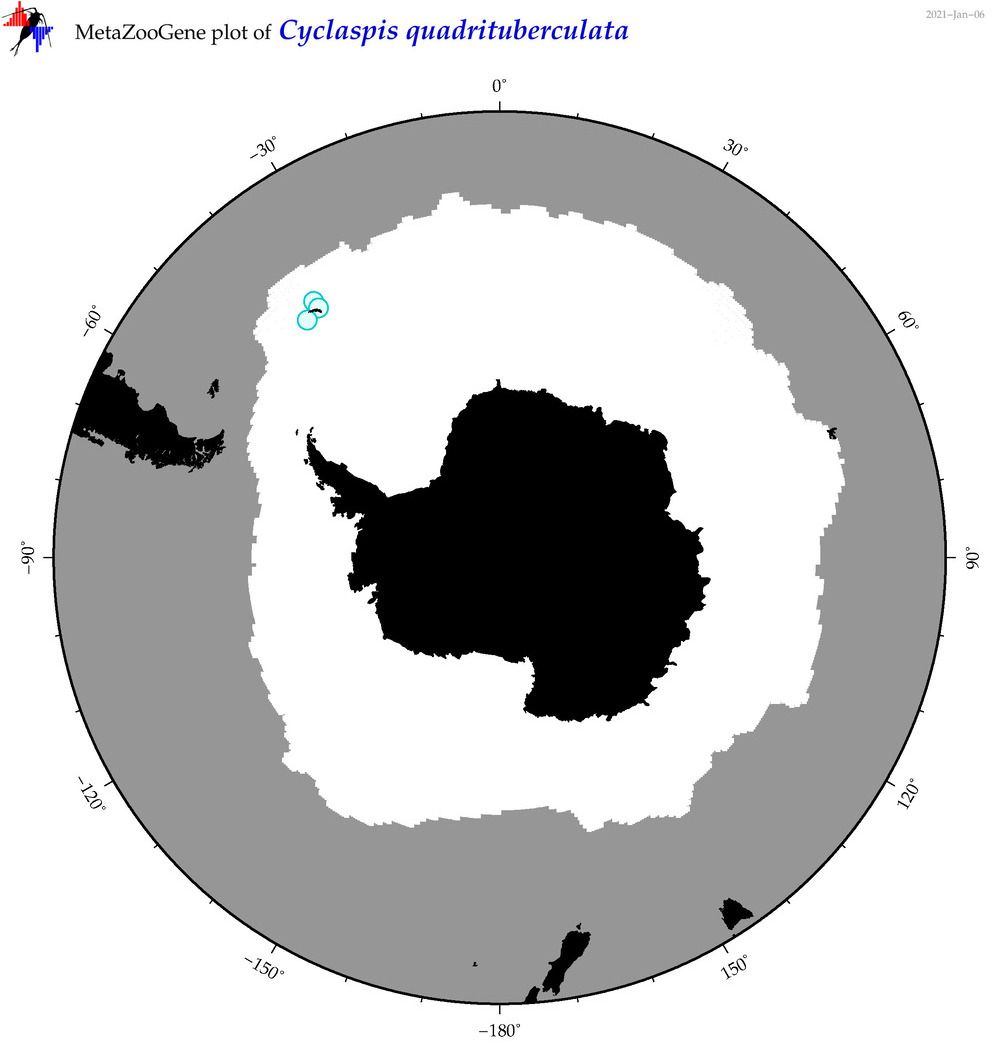 |
accepted
T4048921
R:1:0:0:0 |
| Diastylis anderssoni |
Species
(23) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
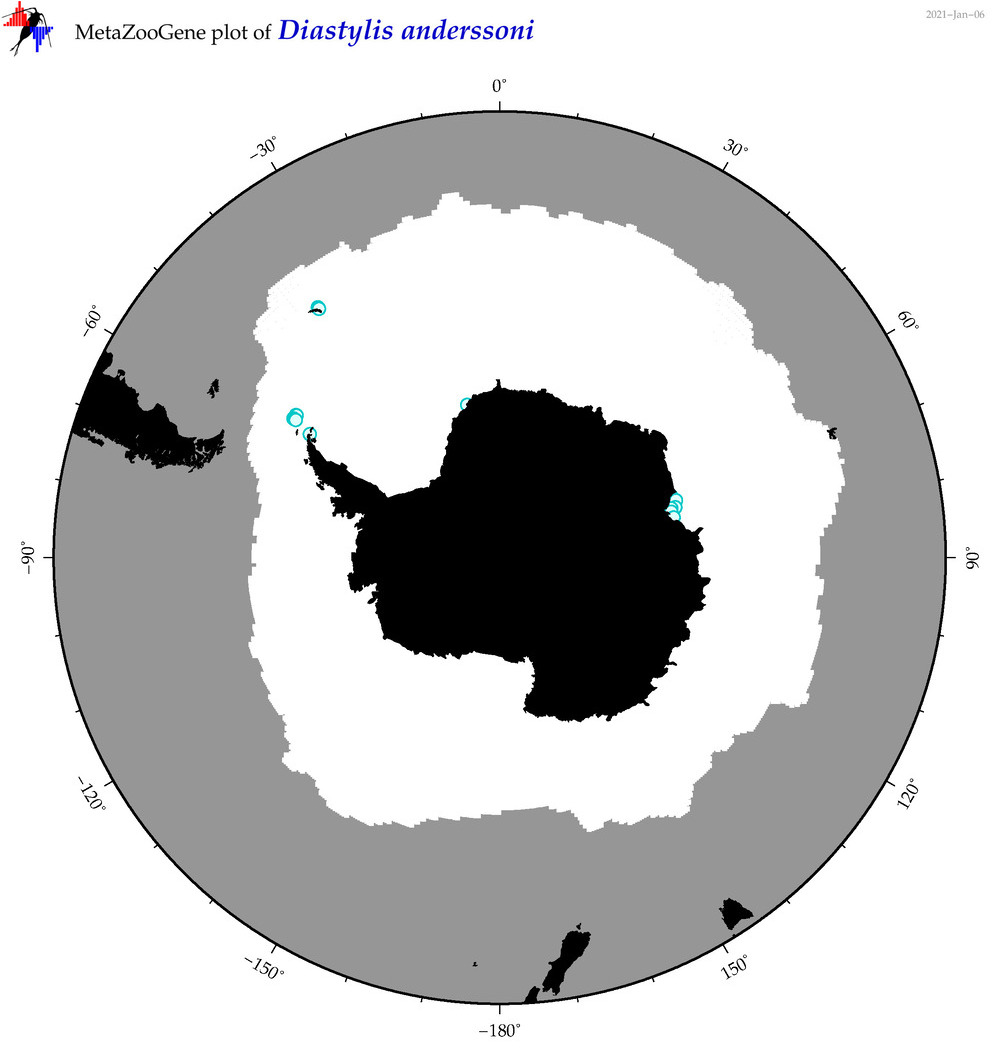 |
accepted
T4049573
R:1:1:0:0 |
| Diastylis corniculata |
Species
(24) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
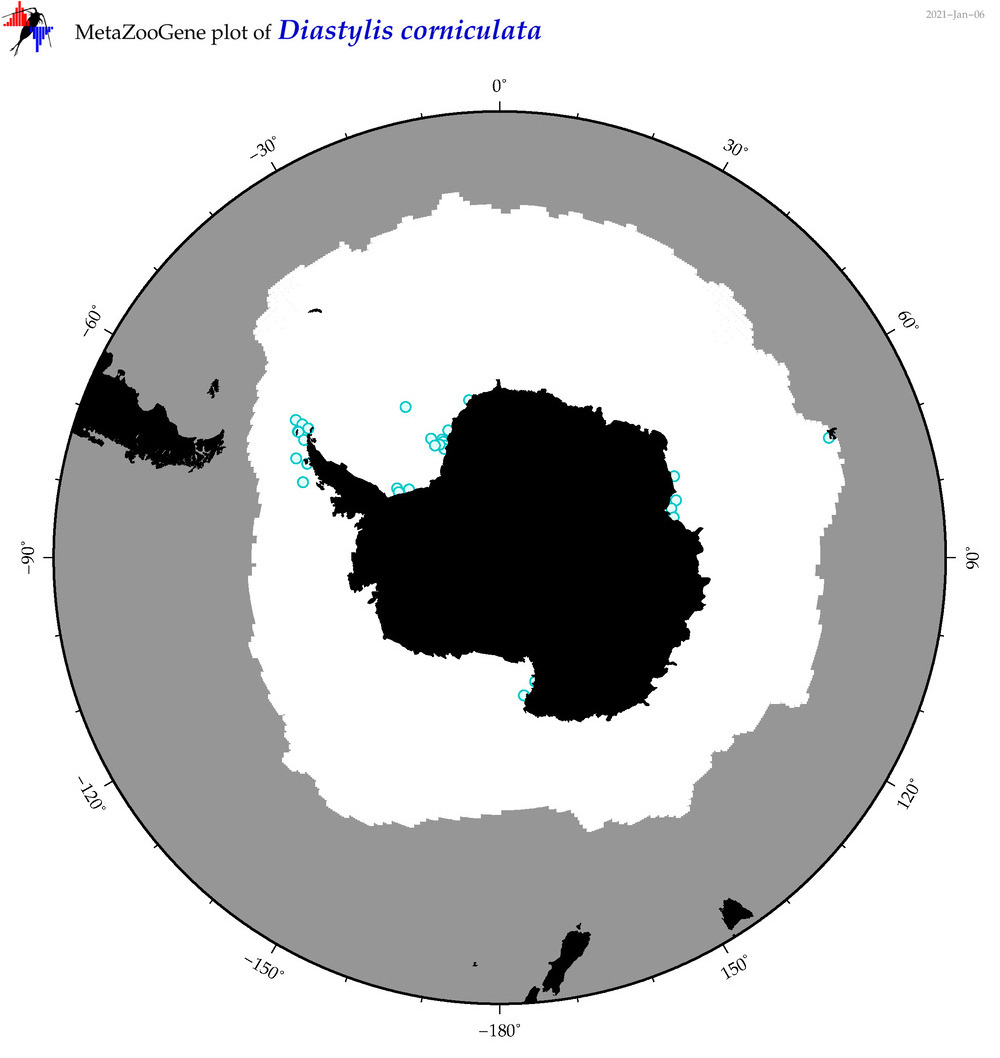 |
accepted
T4049578
R:1:1:0:0 |
| Diastylis enigmatica |
Species
(25) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
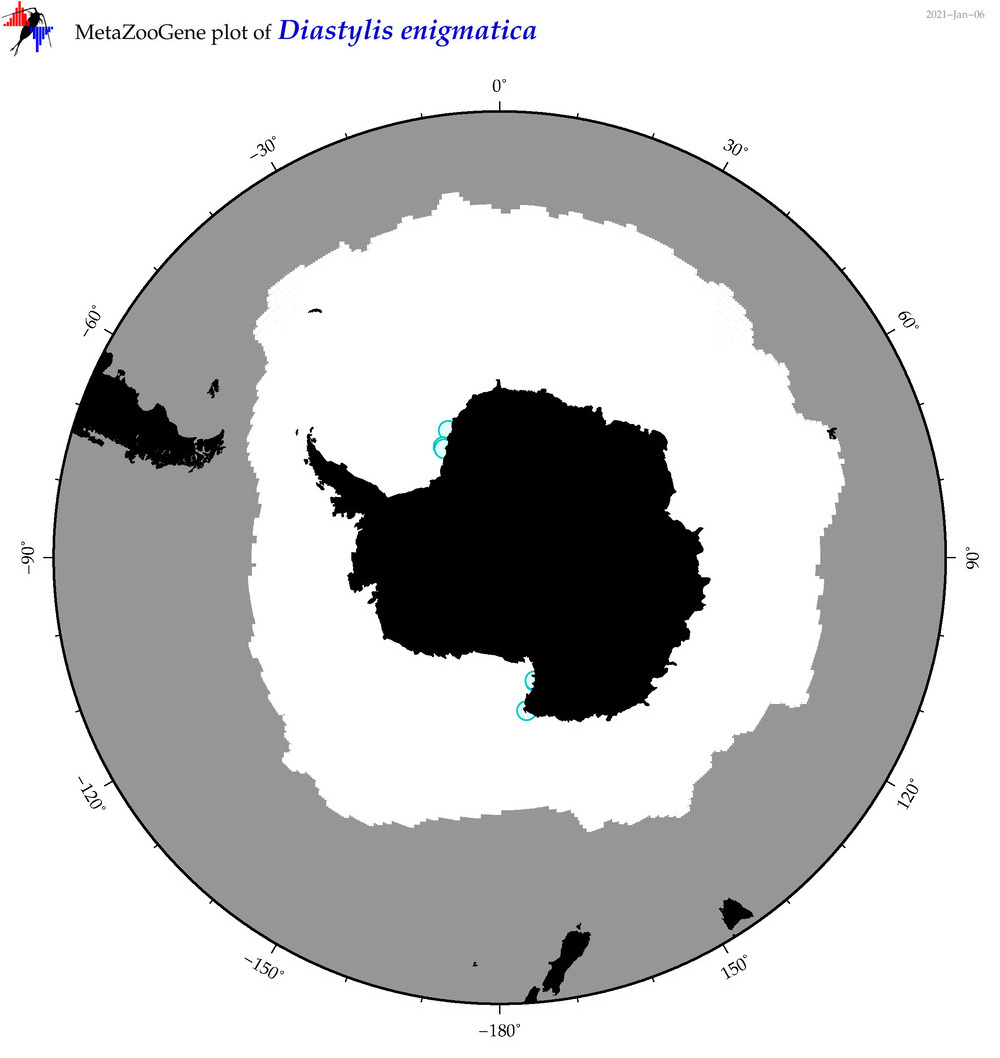 |
accepted
T4049586
R:1:1:0:0 |
| Diastylis galeronae |
Species
(26) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
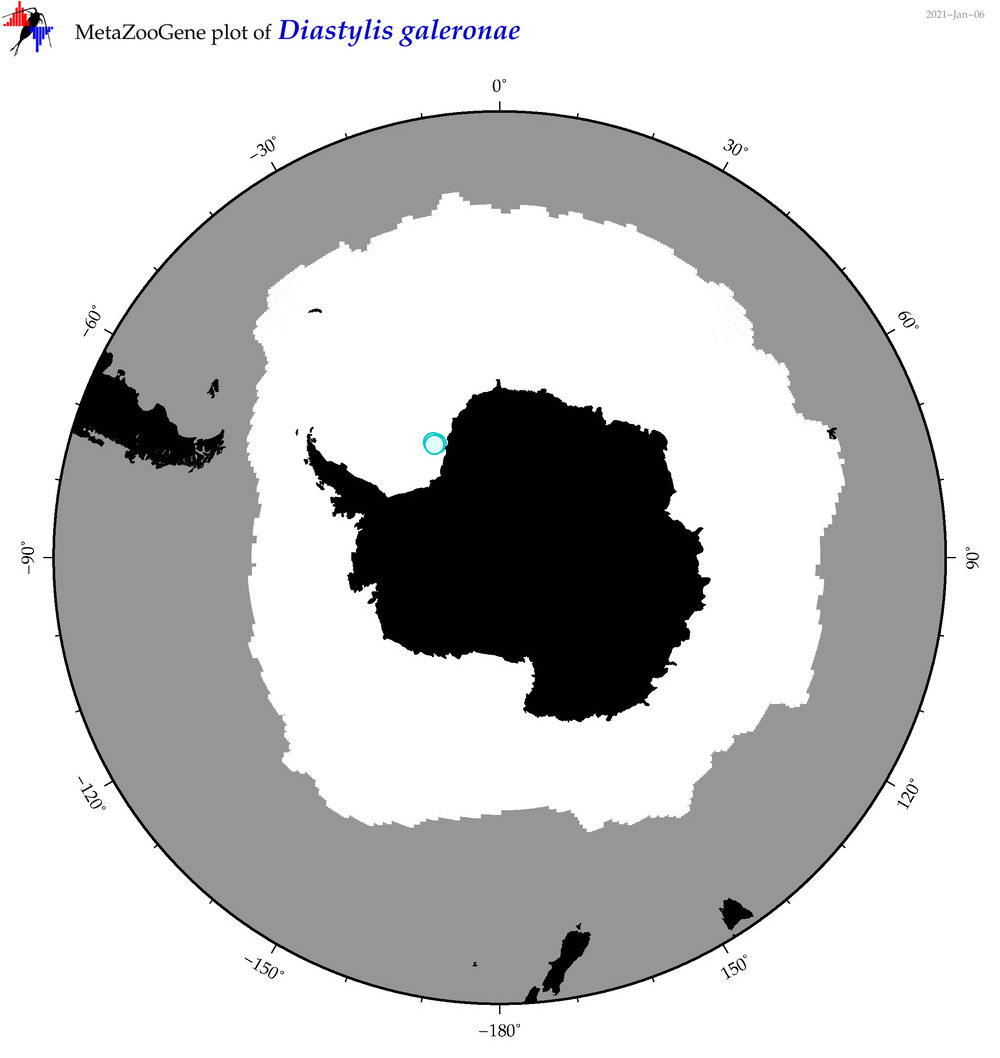 |
accepted
T4049589
R:1:1:0:0 |
| Diastylis horrida |
Species
(27) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4049595
R:1:1:0:0 |
| Diastylis inornata |
Species
(28) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
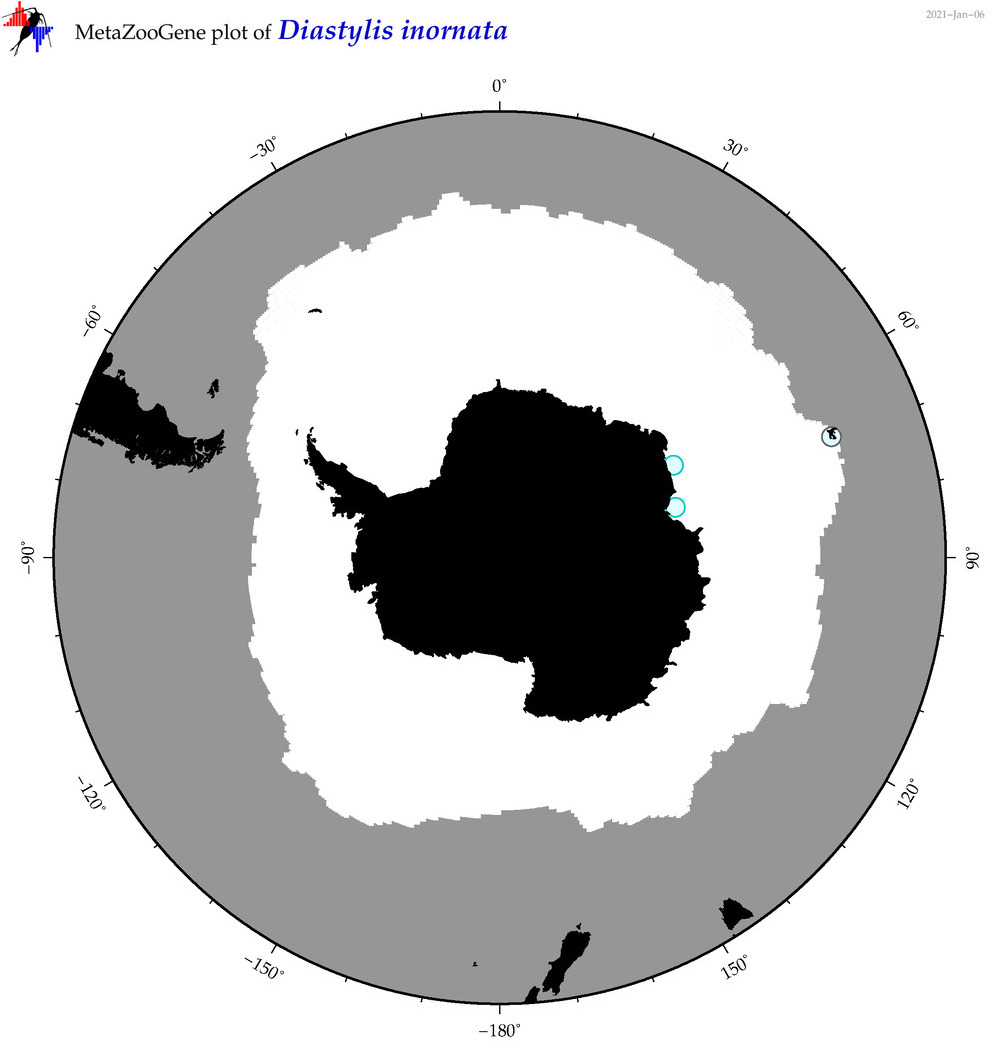 |
accepted
T4049598
R:1:1:0:0 |
| Diastylis mawsoni |
Species
(29) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
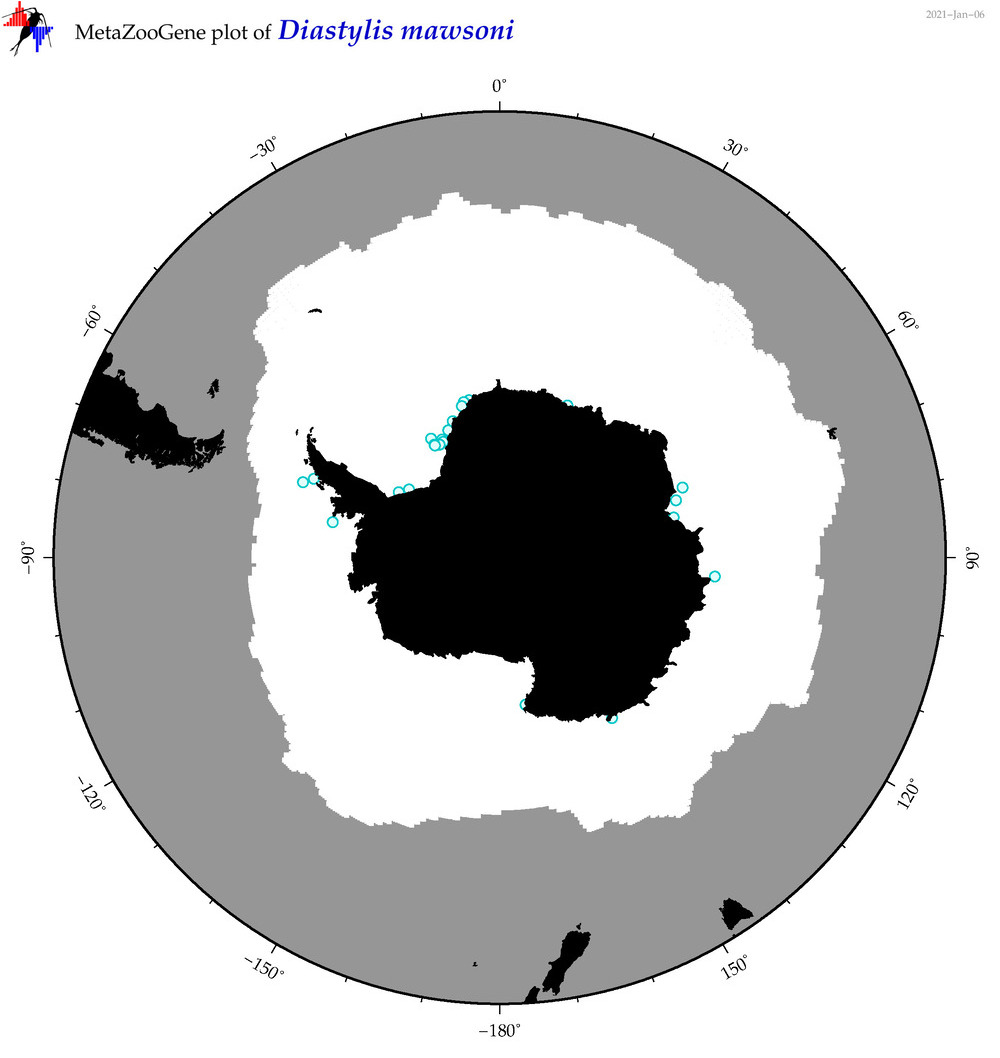 |
accepted
T4049604
R:1:1:0:0 |
| Diastylis planifrons |
Species
(30) |
COI
12S
16S
18S
28S
|
COI = 6
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4049614
R:1:1:0:0 |
| Diastylopsis annulata |
Species
(31) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
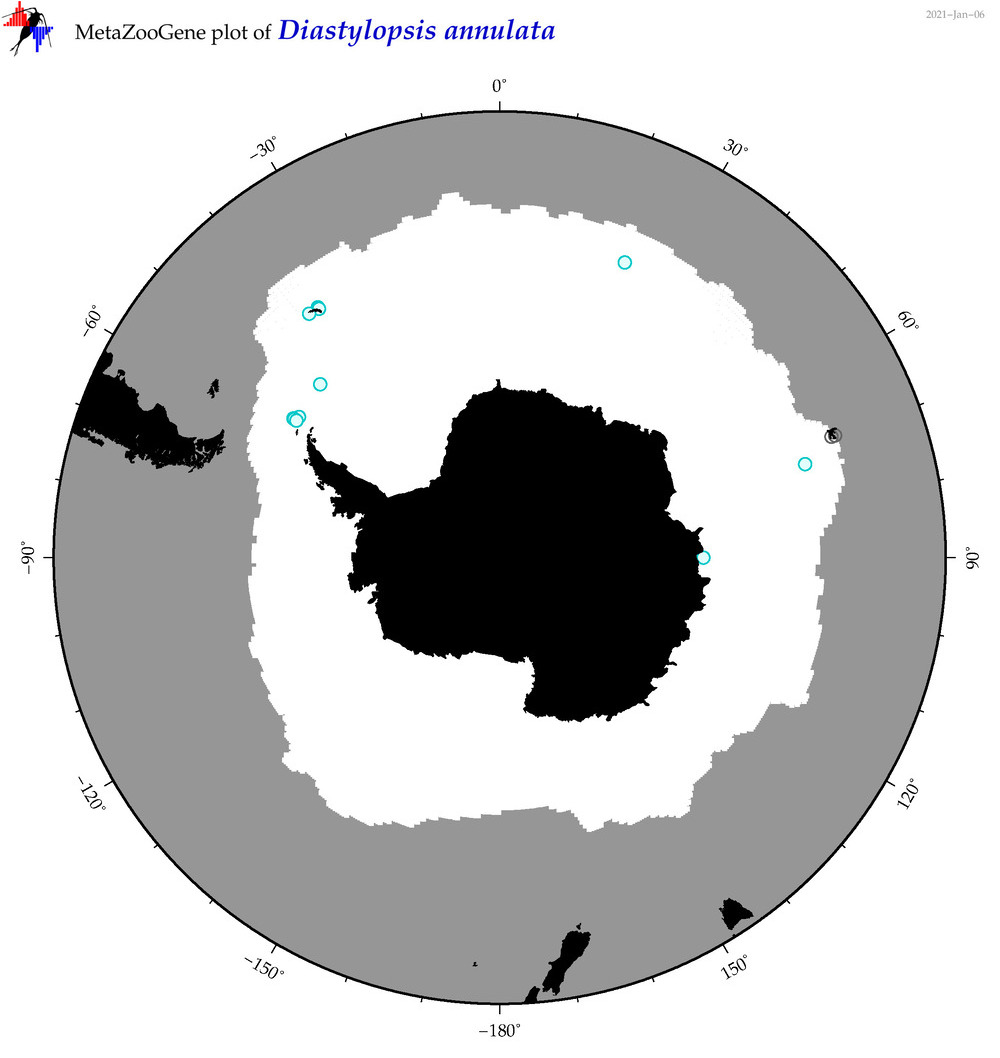 |
accepted
T4049640
R:1:1:0:0 |
| Diastylopsis goekei |
Species
(32) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
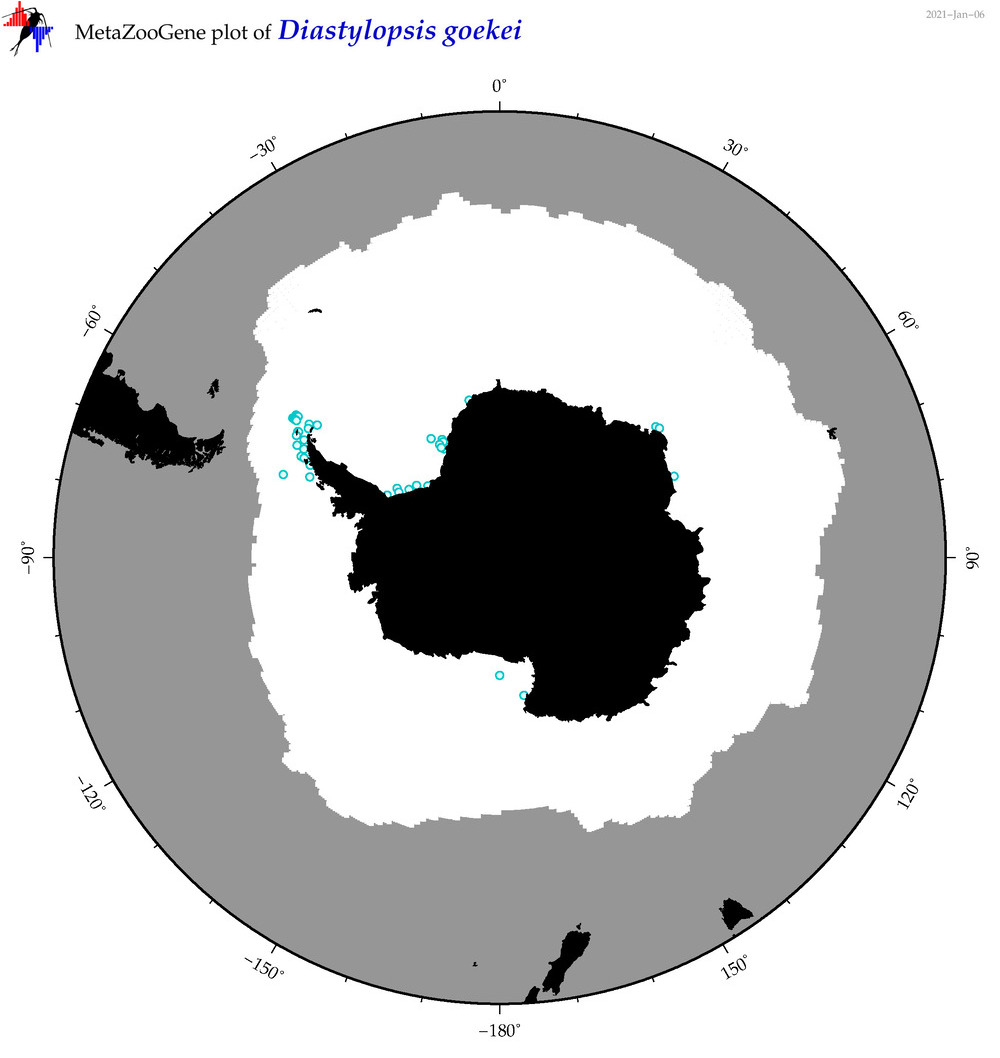 |
accepted
T4049645
R:1:1:0:0 |
| Diastylopsis trisetosa |
Species
(33) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4049648
R:1:1:0:0 |
| Ekleptostylis debroyeri |
Species
(34) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
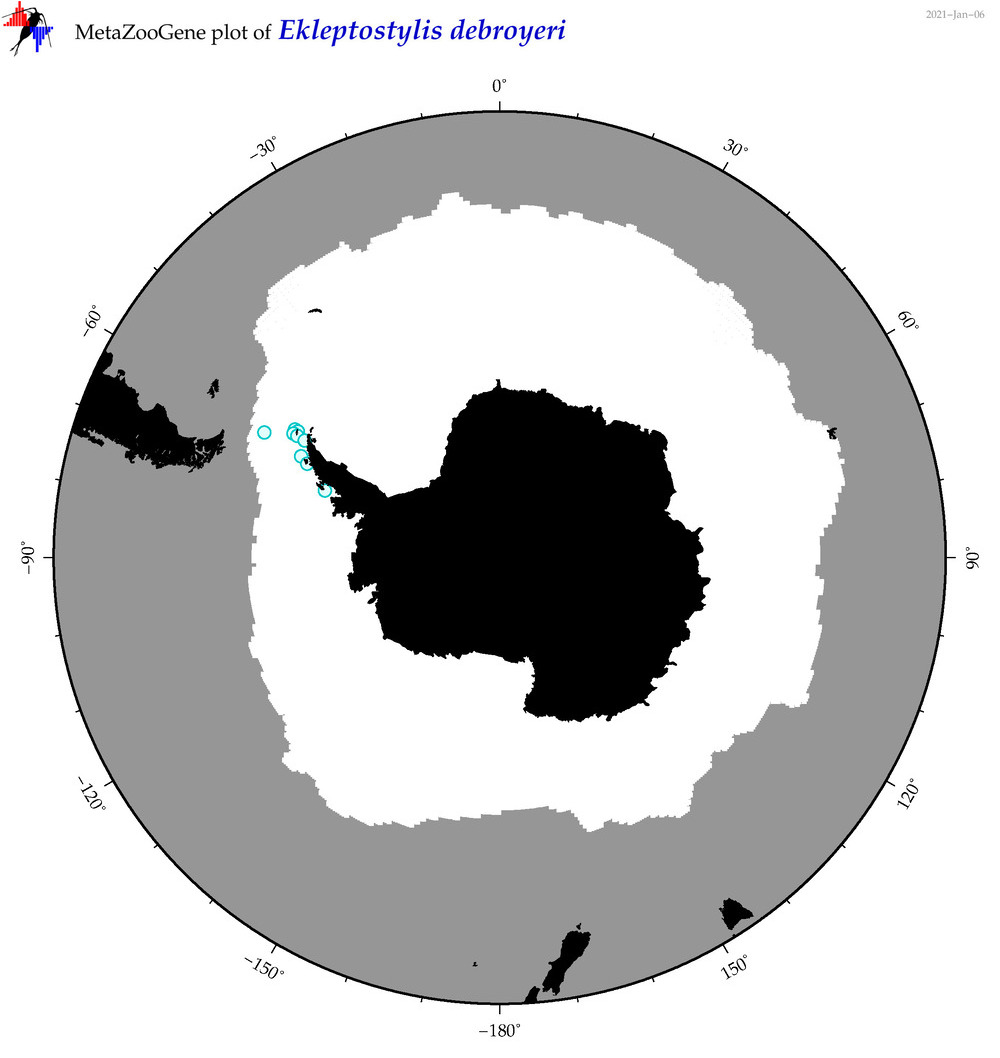 |
accepted
T4050577
R:1:1:0:0 |
| Eudorella fallax |
Species
(35) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
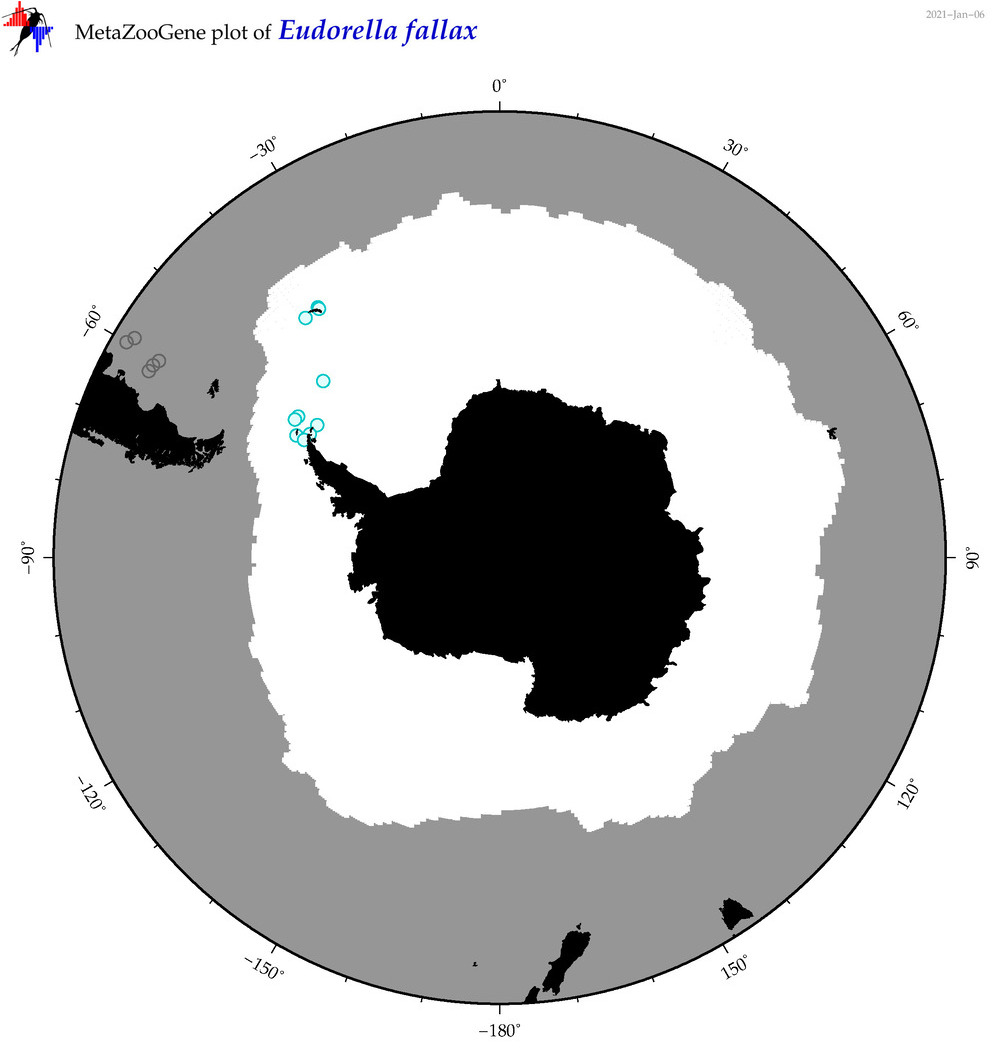 |
accepted
T4051388
R:1:1:0:0 |
| Eudorella gracilior |
Species
(36) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
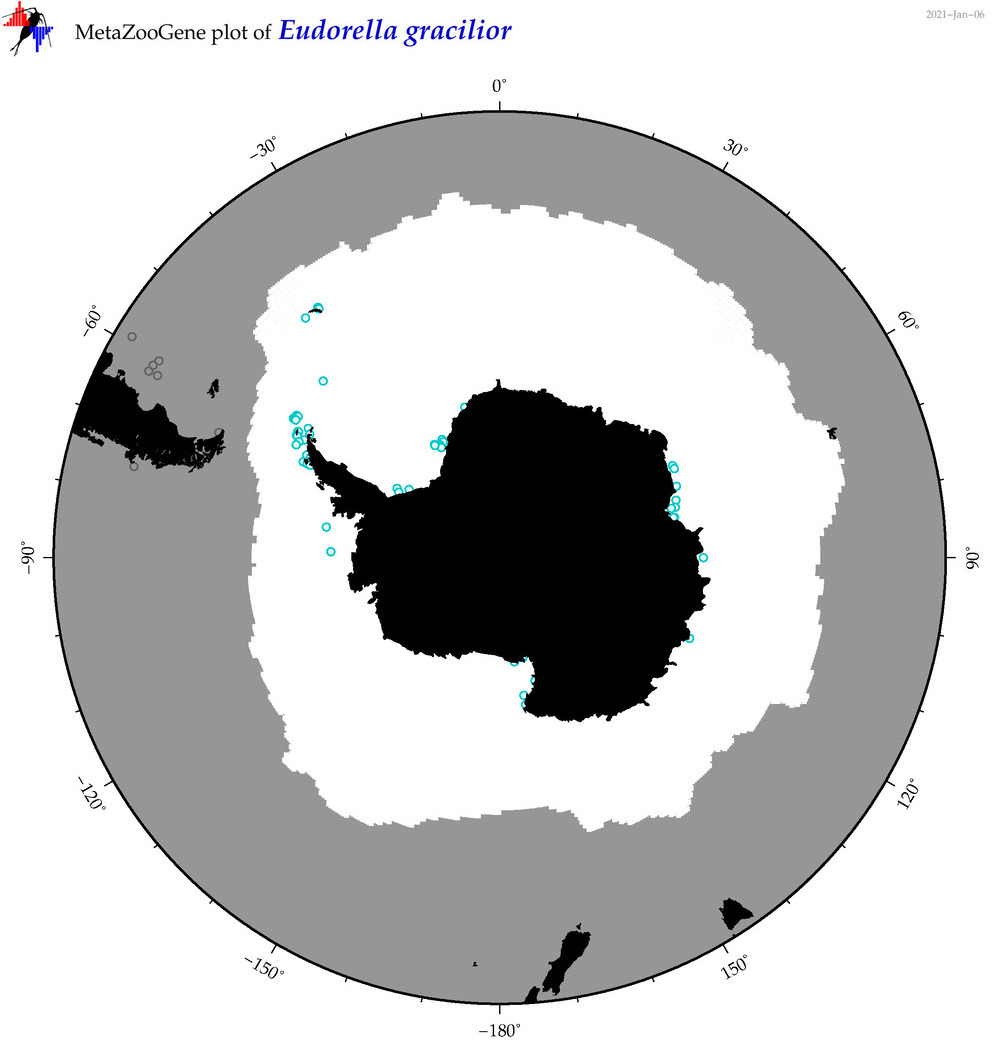 |
accepted
T4051389
R:1:1:0:0 |
| Eudorella similis |
Species
(37) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
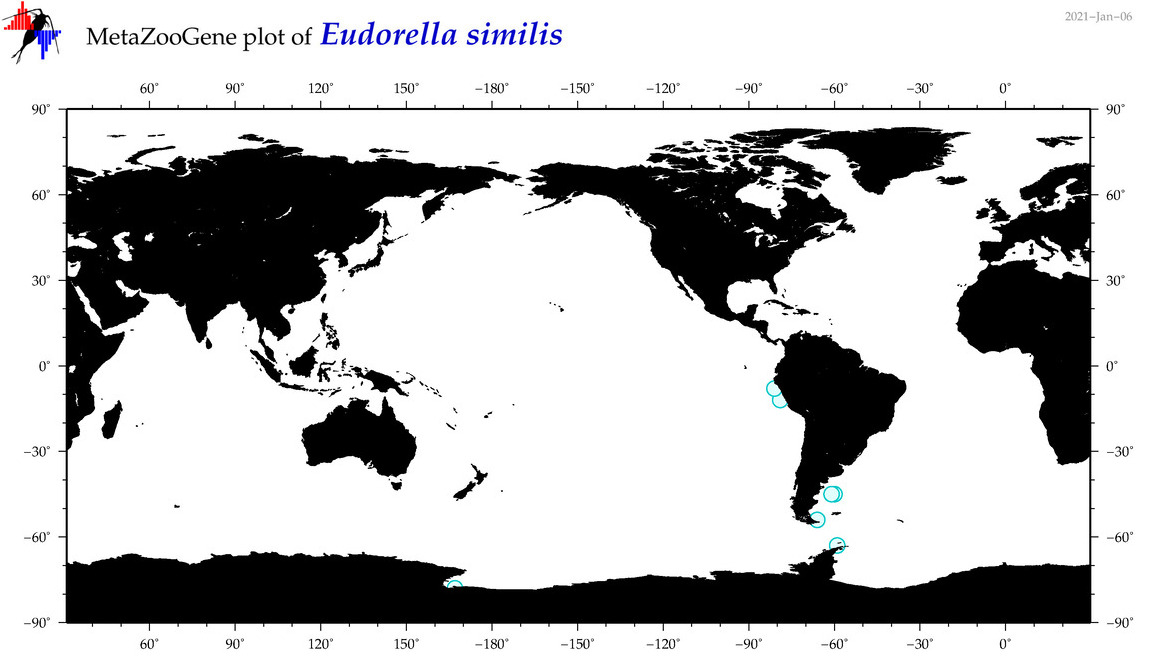 |
accepted
T4051400
R:1:1:0:0 |
| Eudorella sordida |
Species
(38) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
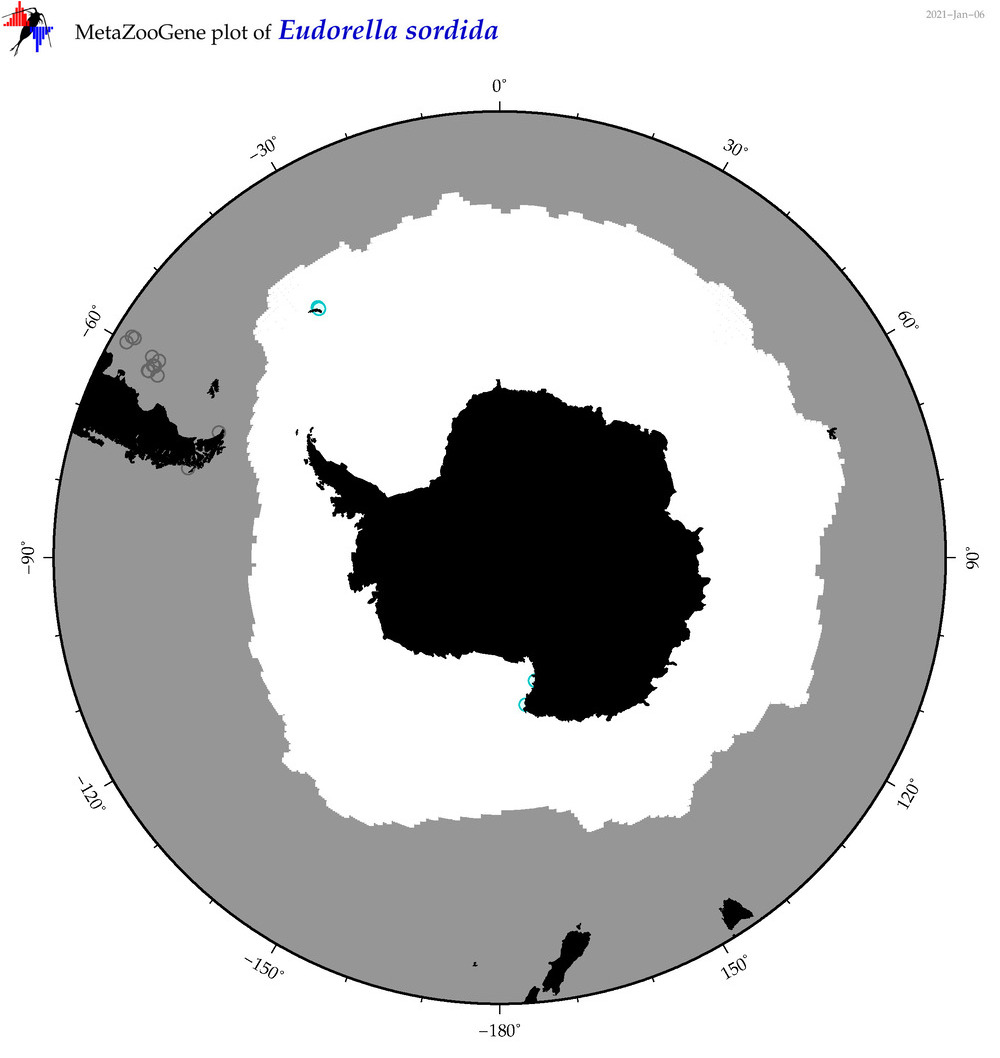 |
accepted
T4051401
R:1:1:0:0 |
| Eudorella splendida |
Species
(39) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
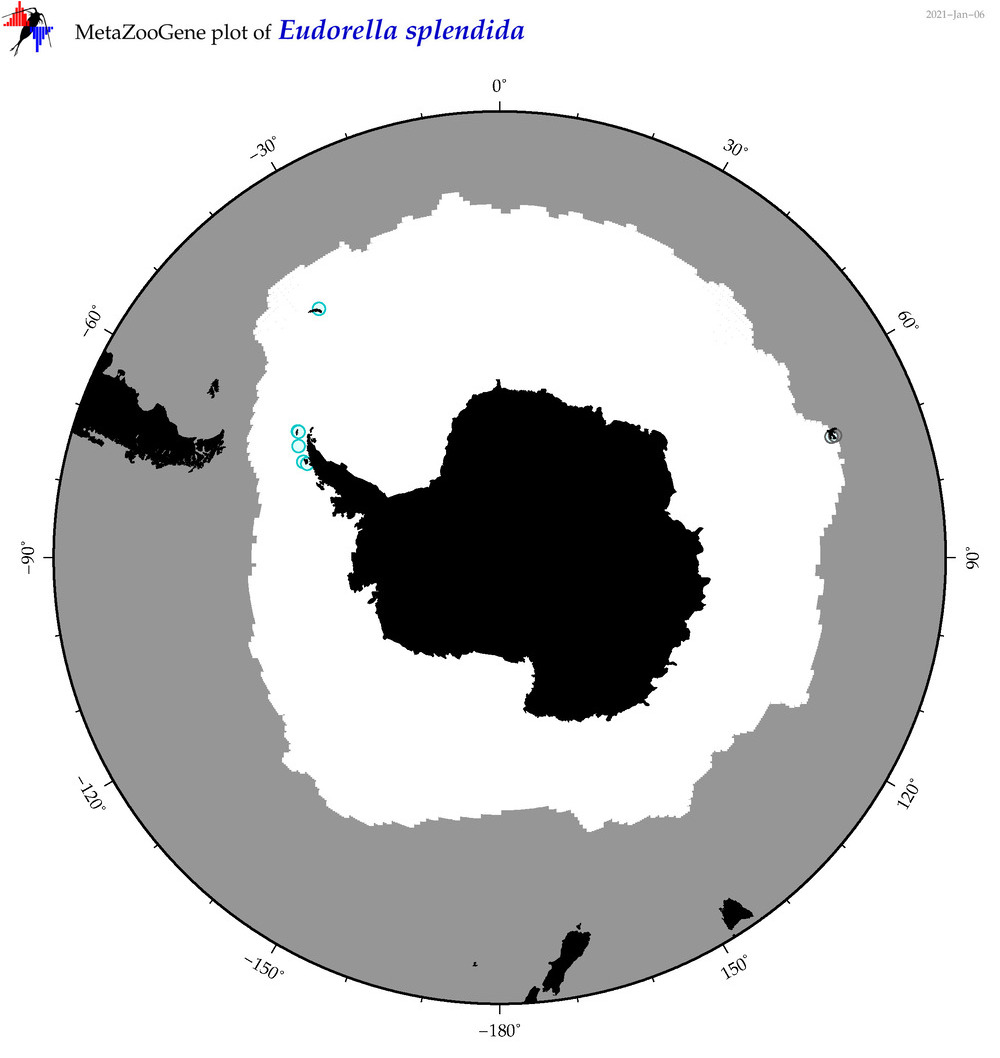 |
accepted
T4051403
R:1:1:0:0 |
| Eudorella truncatula |
Species
(40) |
COI
12S
16S
18S
28S
|
COI = 14
12S = 0
16S = 0
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
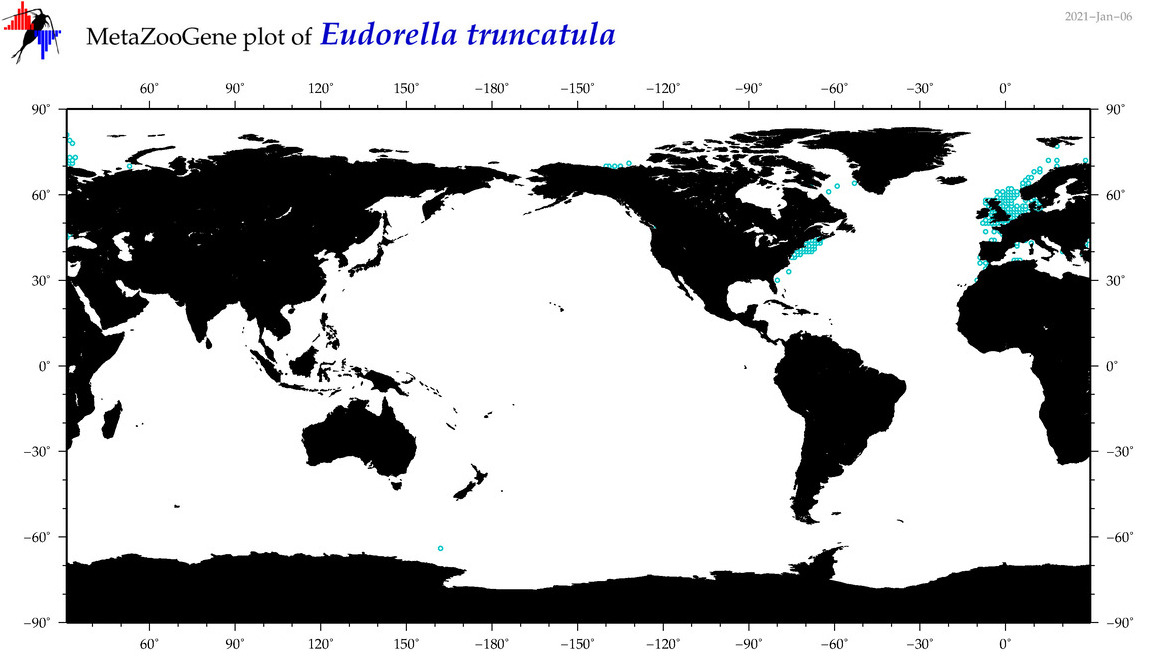 |
accepted
T4051404
R:1:1:0:0 |
| Gaussicuma vanhoeffeni |
Species
(41) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
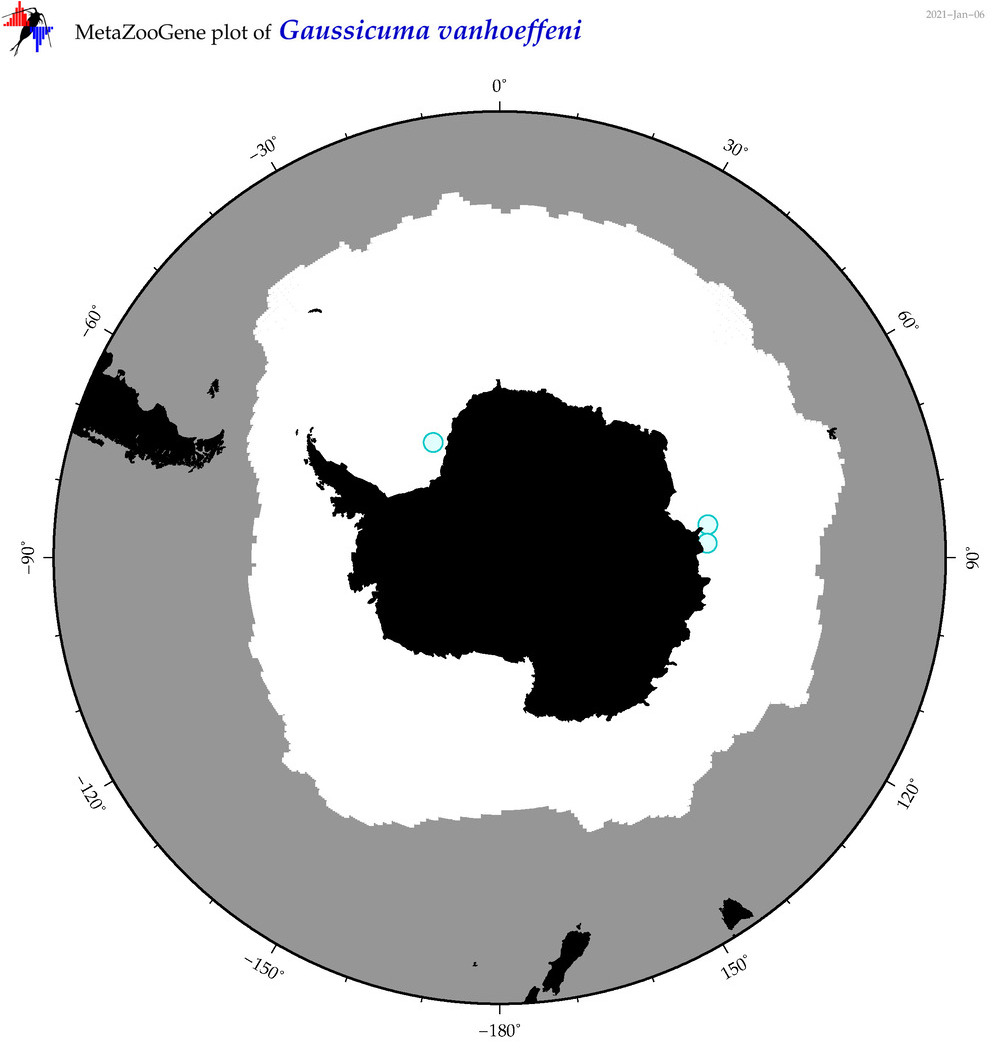 |
accepted
T4052615
R:1:0:0:0 |
| Hemilamprops bacescui |
Species
(42) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
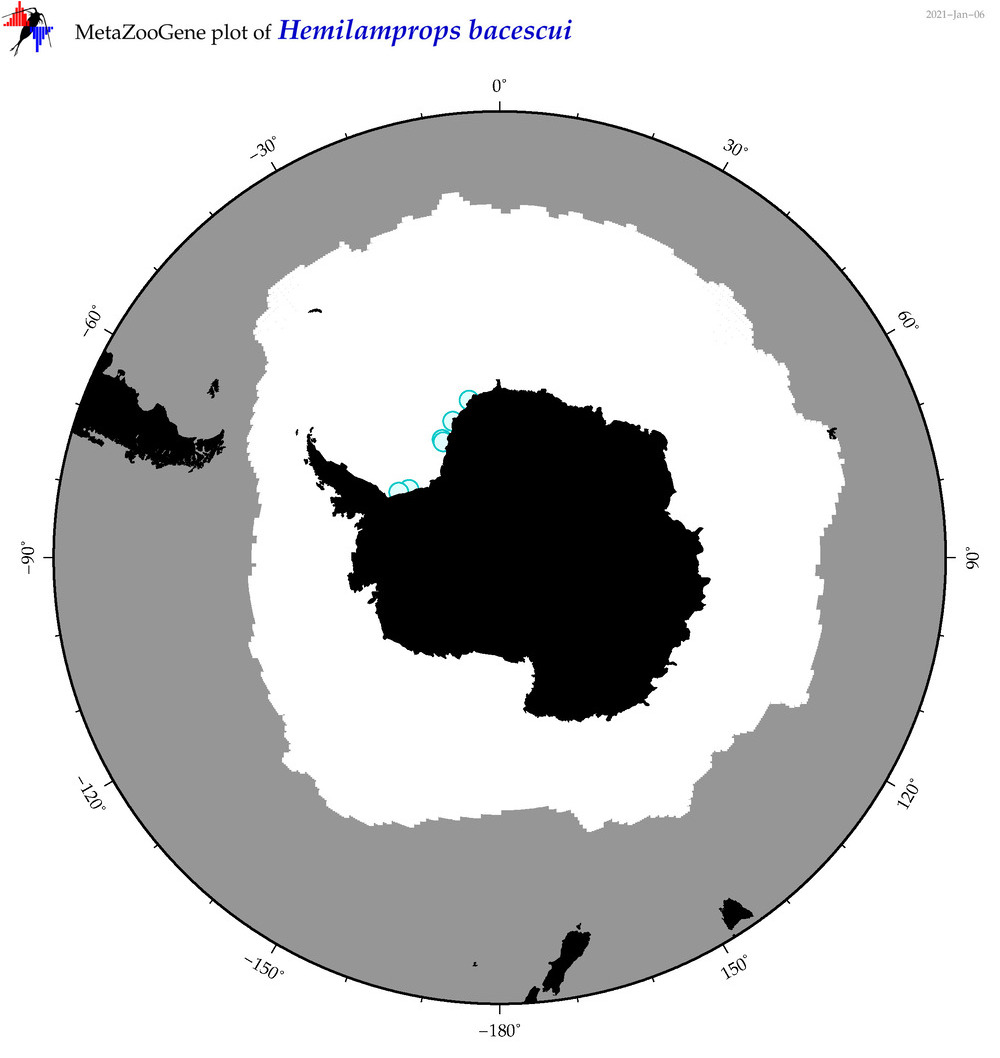 |
accepted
T4053842
R:1:0:0:0 |
| Hemilamprops pellucidus |
Species
(43) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
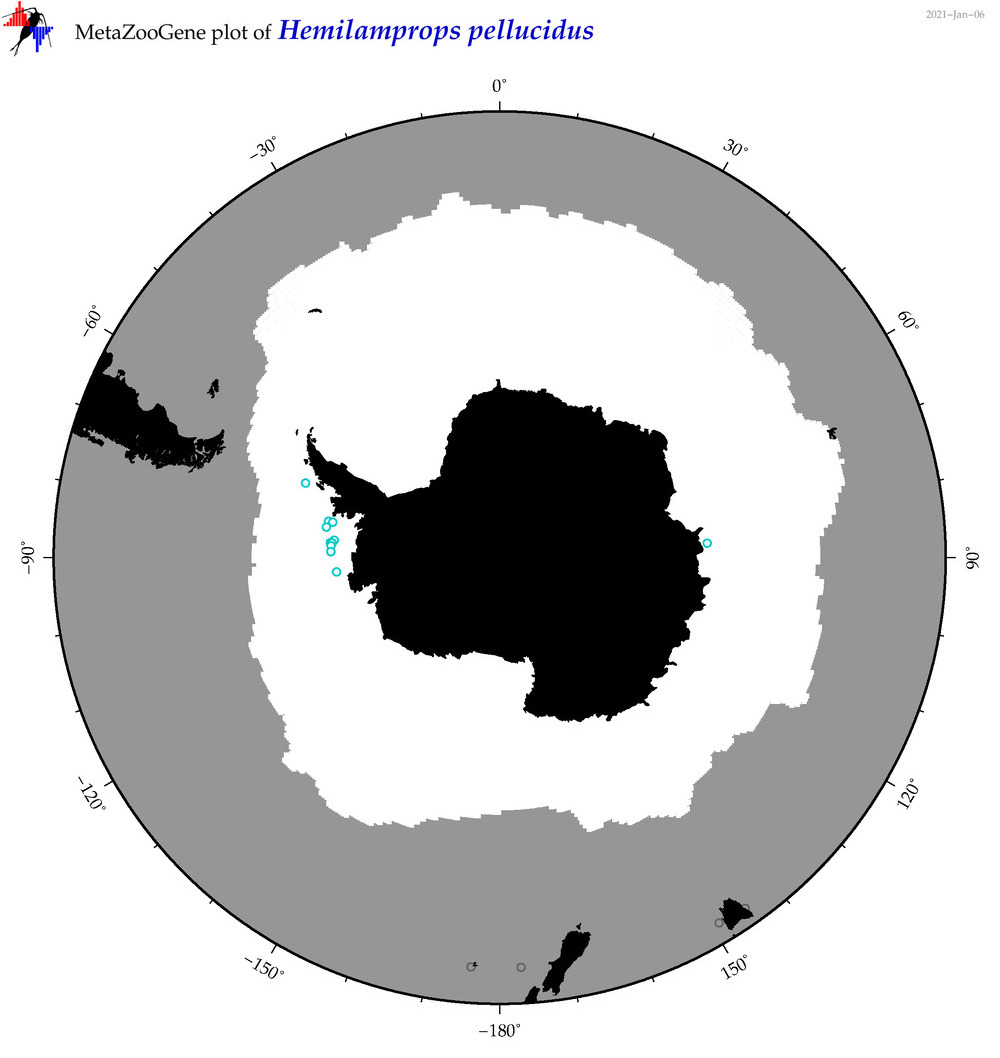 |
accepted
T4053851
R:1:0:0:0 |
| Hemilamprops ultimaespei |
Species
(44) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
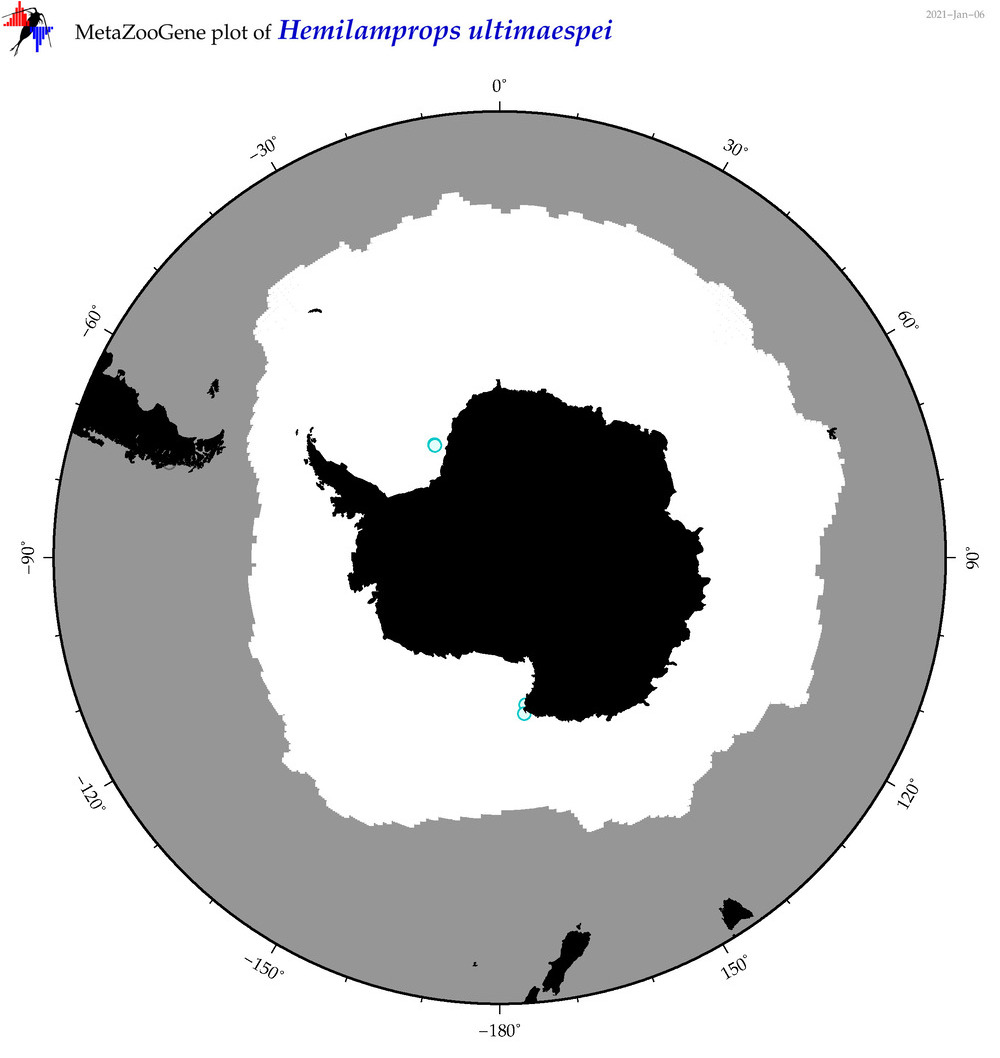 |
accepted
T4053853
R:1:0:0:0 |
| Holostylis helleri |
Species
(45) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
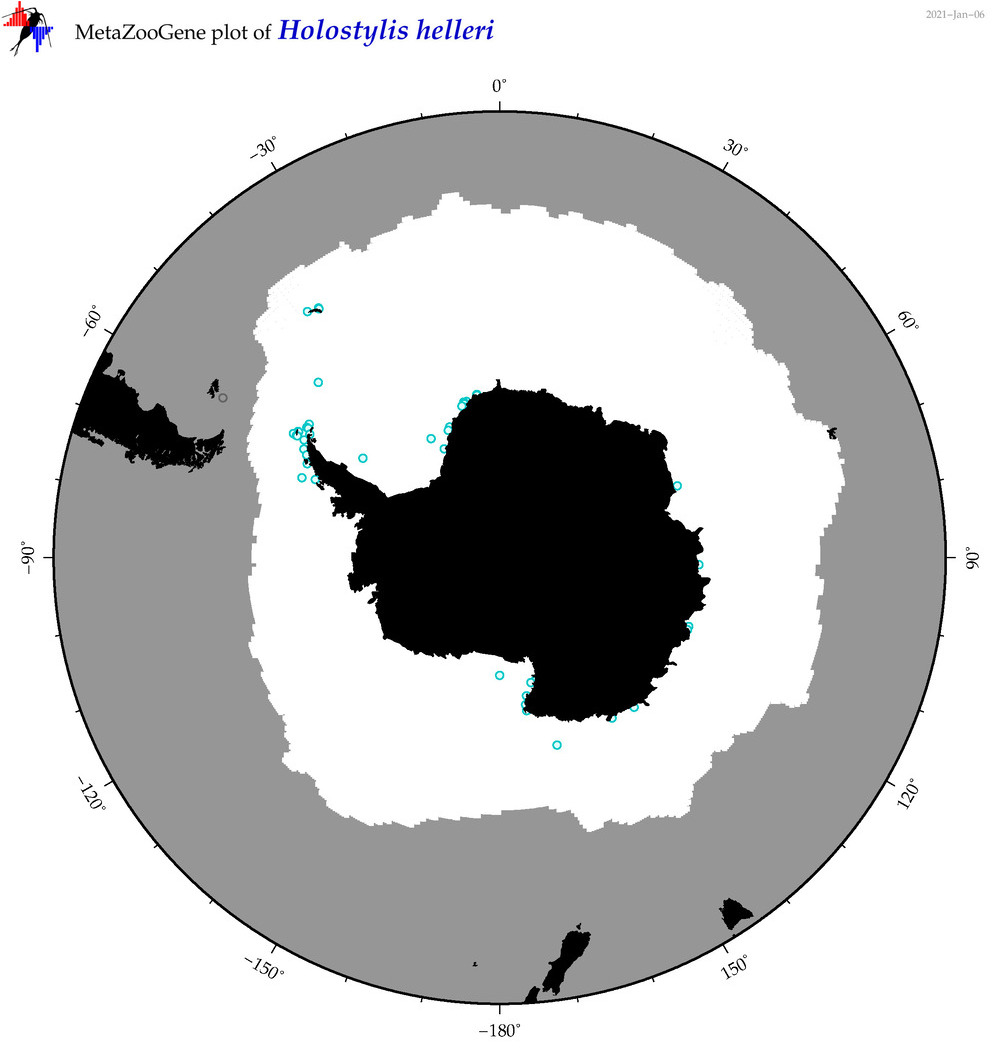 |
accepted
T4054319
R:1:1:0:0 |
| Holostylis spinicauda |
Species
(46) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4054320
R:1:1:0:0 |
| Leptostylis antipus |
Species
(47) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
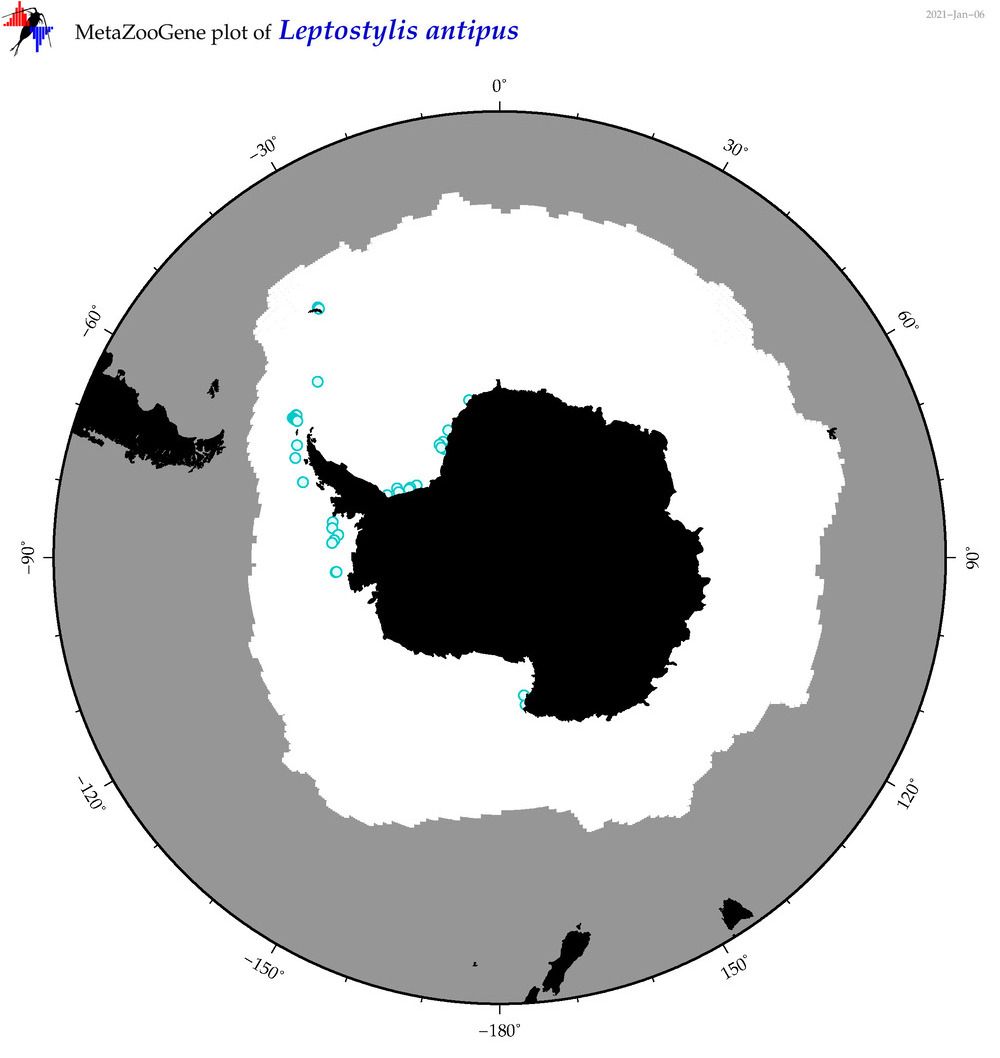 |
accepted
T4056229
R:1:1:0:0 |
| Leptostylis crassicauda |
Species
(48) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
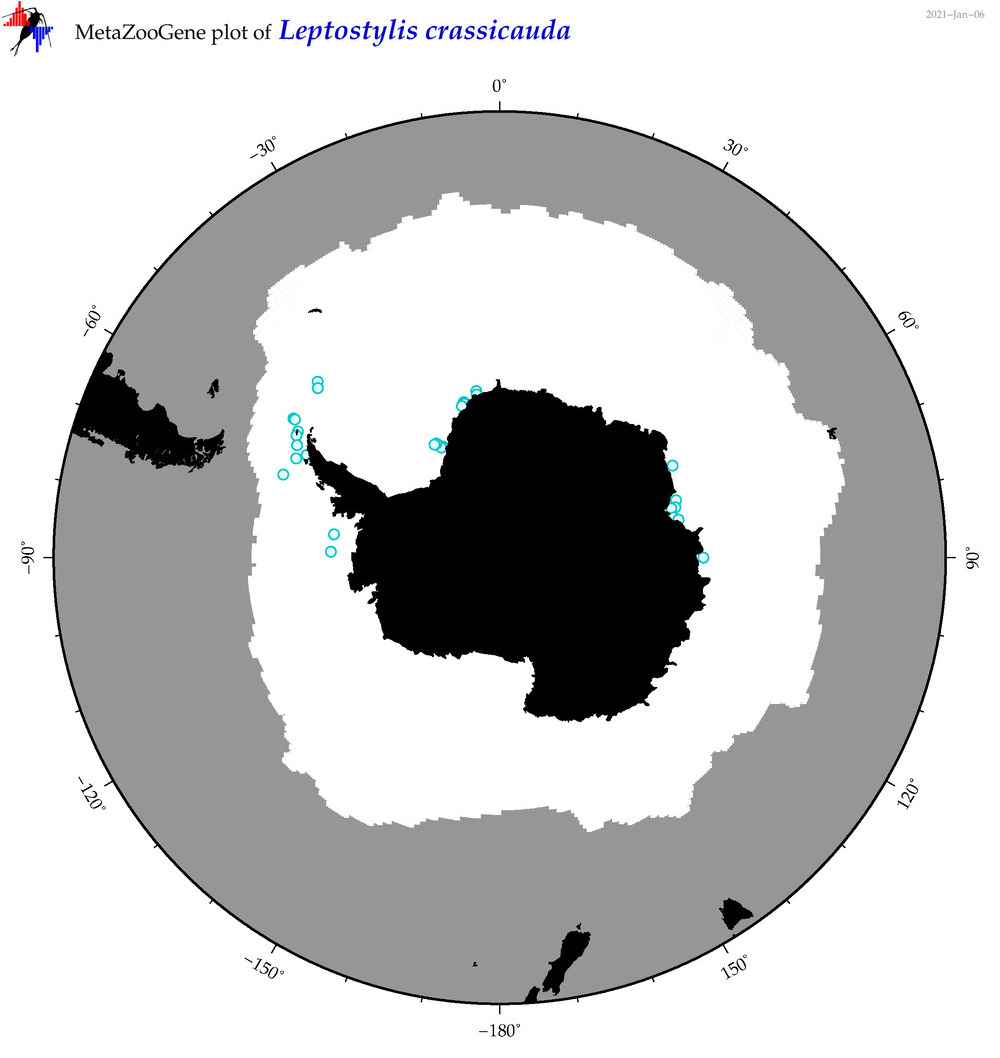 |
accepted
T4056235
R:1:1:0:0 |
| Leptostylis weddelli |
Species
(49) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
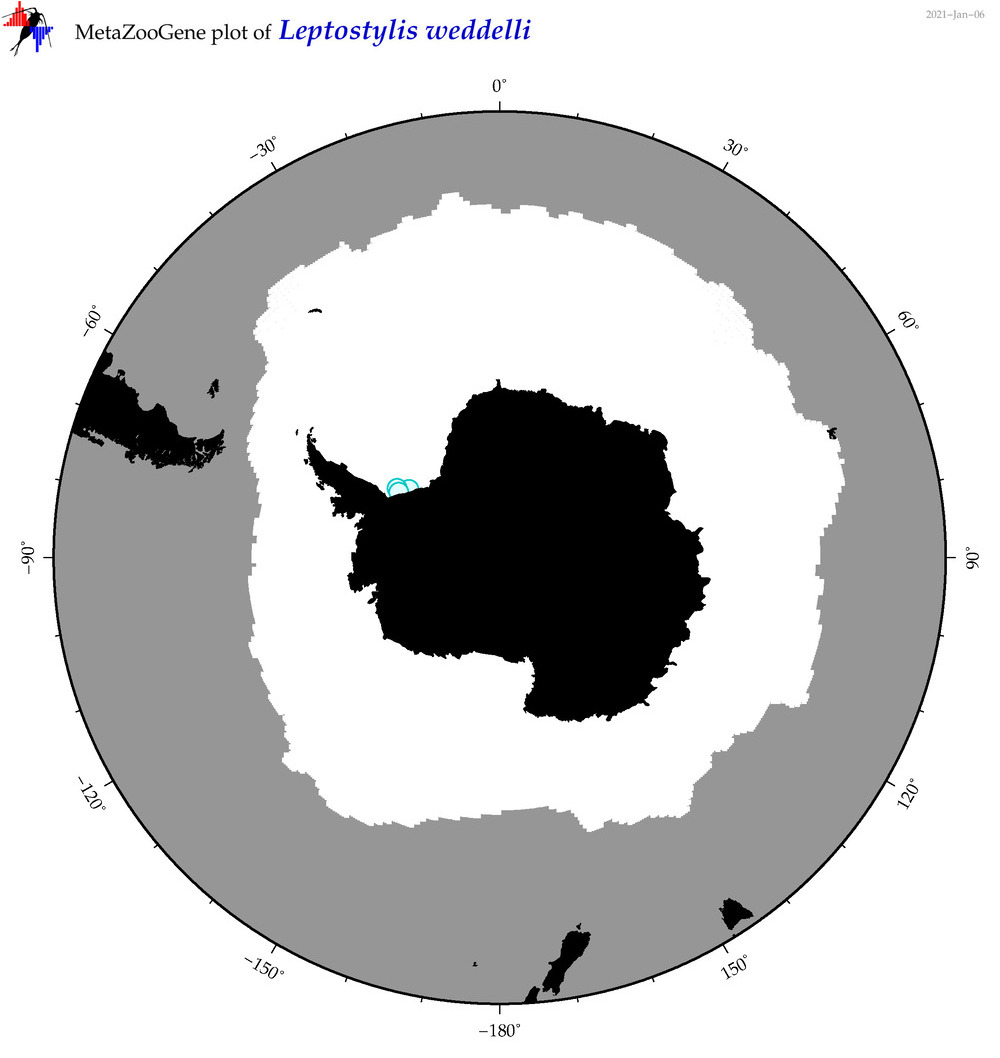 |
accepted
T4056254
R:1:1:0:0 |
| Leucon (Alytoleucon) polarsterni |
Species
(50) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4094838
R:1:1:0:0 |
| Leucon (Crymoleucon) antarcticus |
Species
(51) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
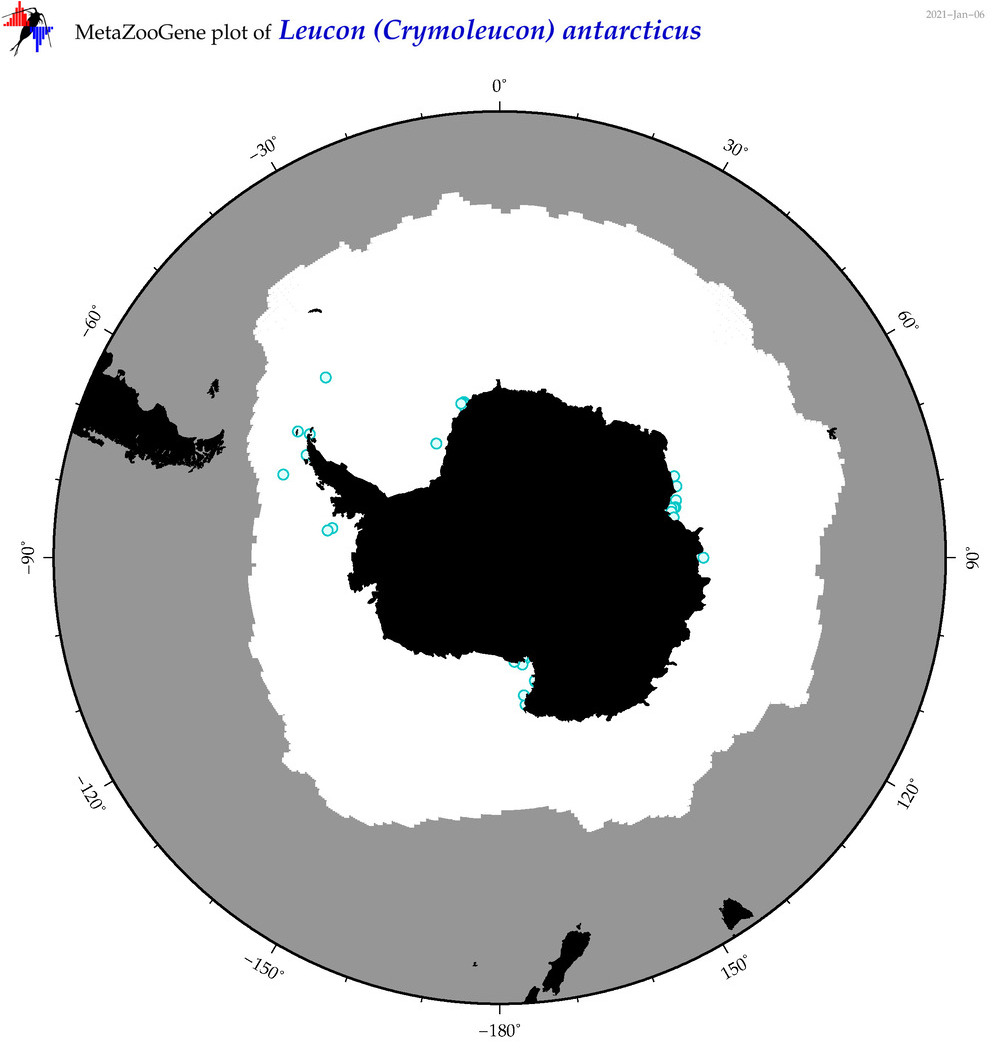 |
accepted
T4094840
R:1:1:0:0 |
| Leucon (Crymoleucon) breidensis |
Species
(52) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4131216
R:1:1:0:0 |
| Leucon (Crymoleucon) costatus |
Species
(53) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4131217
R:1:1:0:0 |
| Leucon (Crymoleucon) intermedius |
Species
(54) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 8
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 8
18S = 0
28S = 0
|
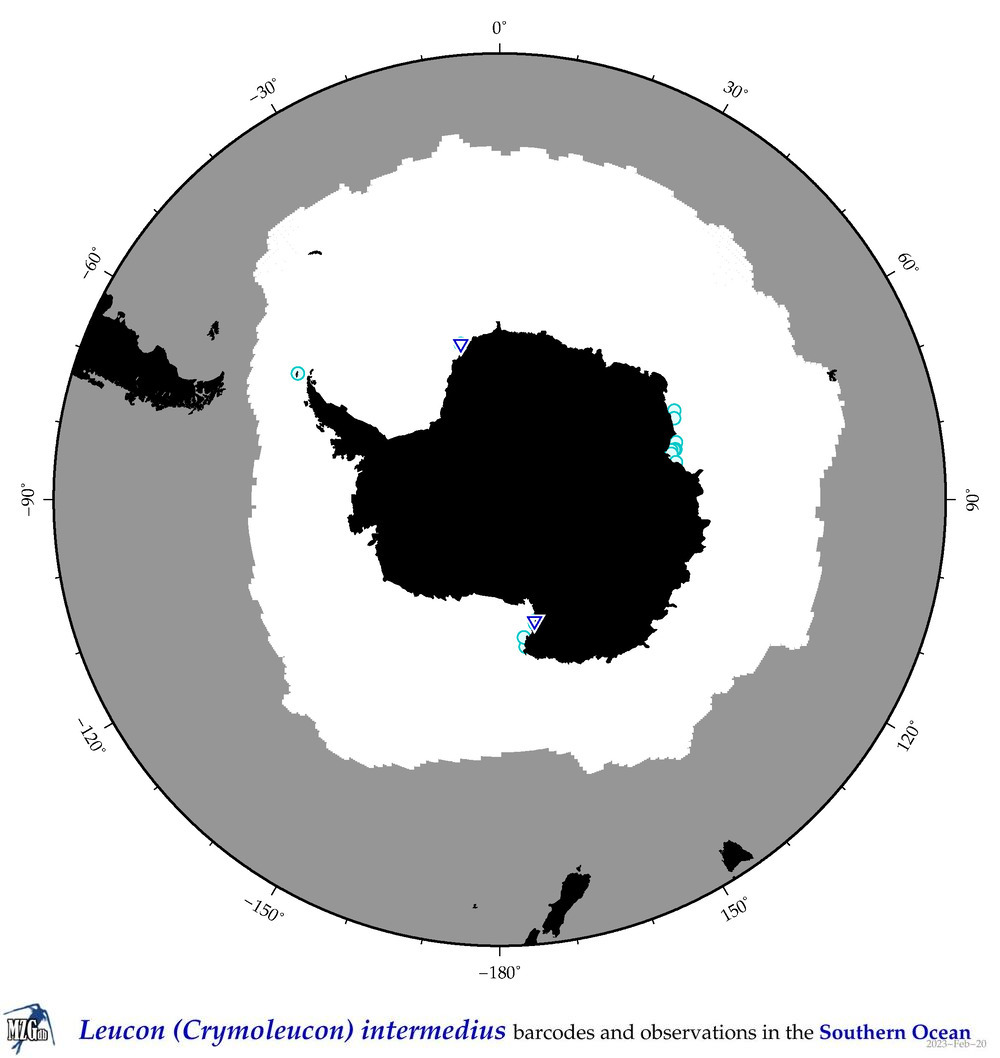 |
accepted
T4131218
R:1:1:0:0 |
| Leucon (Crymoleucon) sagitta |
Species
(55) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
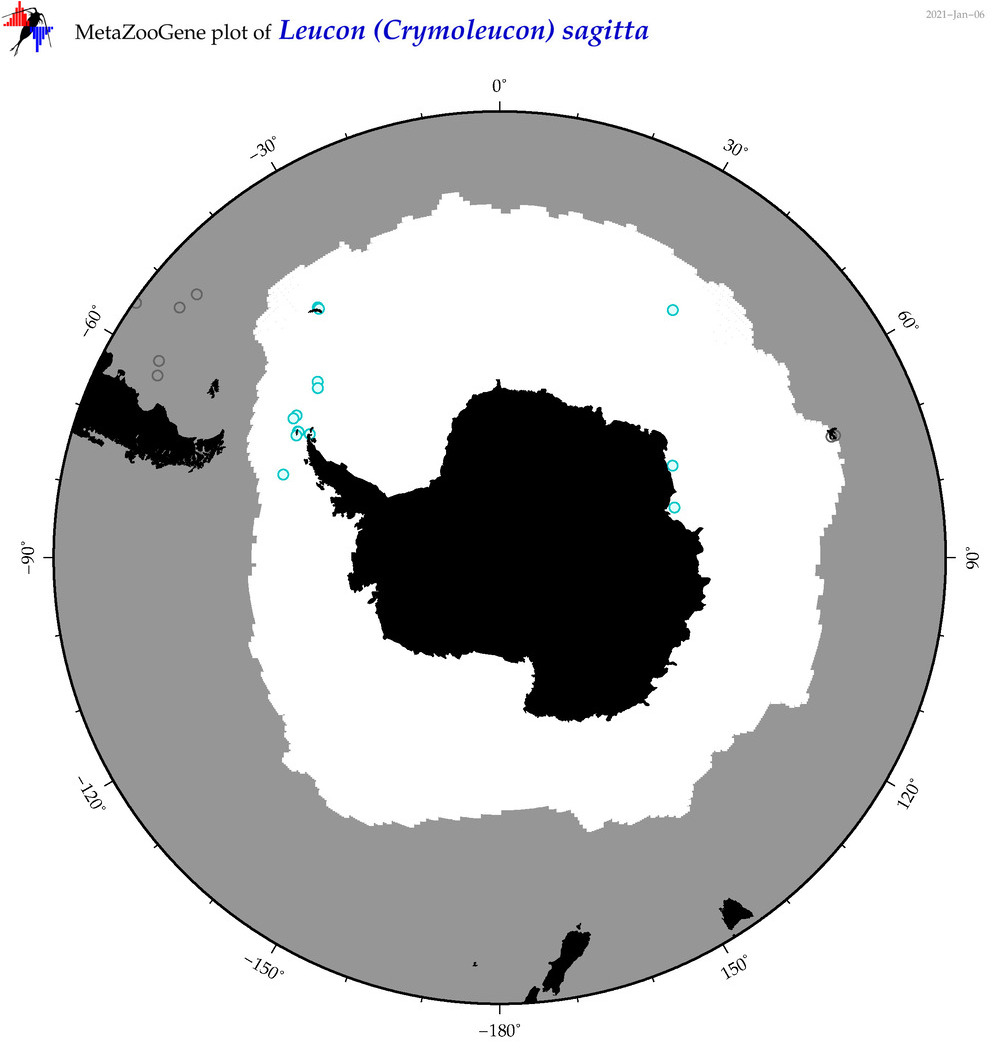 |
accepted
T4094844
R:1:1:0:0 |
| Leucon (Leucon) adelae |
Species
(56) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4094856
R:1:1:0:0 |
| Leucon (Leucon) assimilis |
Species
(57) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4094857
R:1:1:0:0 |
| Leucon (Macrauloleucon) parasiphonatus |
Species
(58) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4131220
R:1:1:0:0 |
| Leucon (Macrauloleucon) weddelli |
Species
(59) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
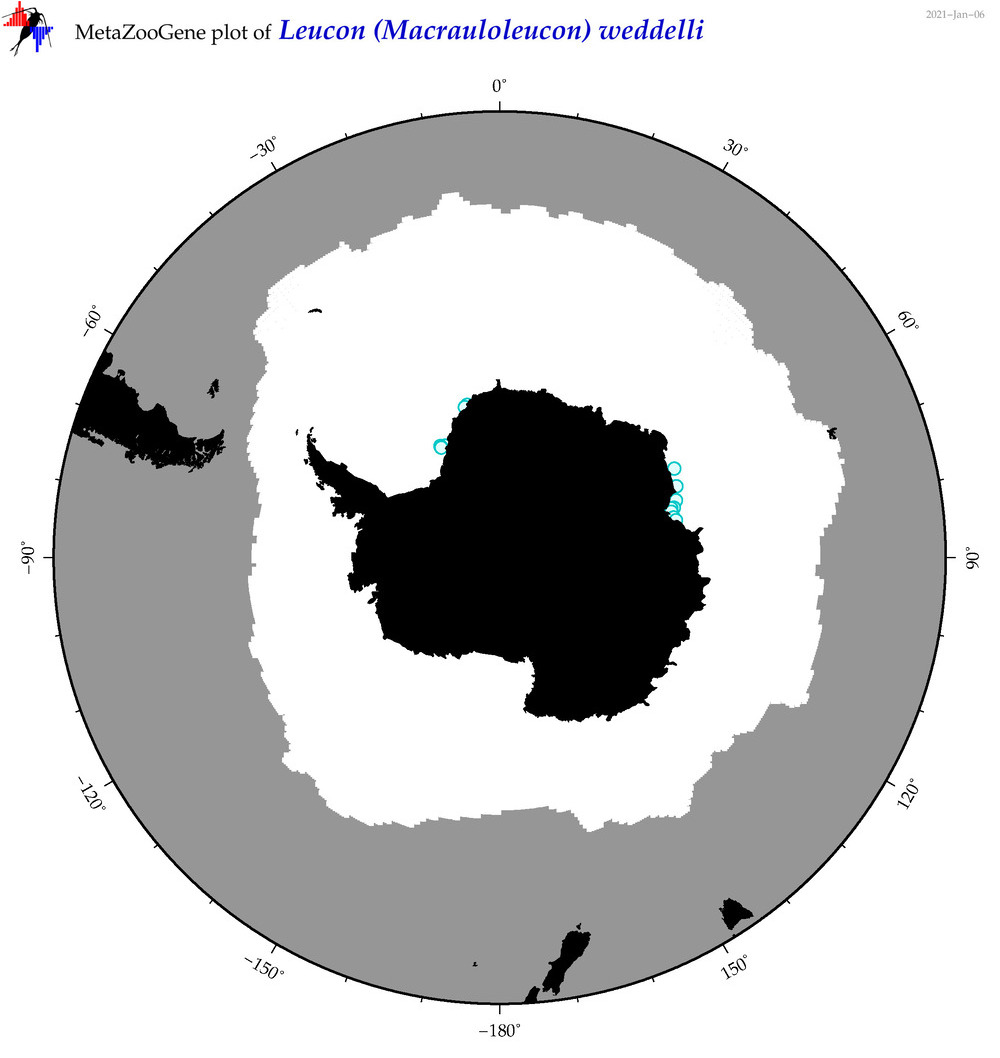 |
accepted
T4094869
R:1:1:0:0 |
| Litogynodiastylis diaphanes |
Species
(60) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4056668
R:1:0:0:0 |
| Makrokylindrus (Adiastylis) baceskei |
Species
(61) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4094808
R:1:1:0:0 |
| Makrokylindrus (Adiastylis) inscriptus |
Species
(62) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
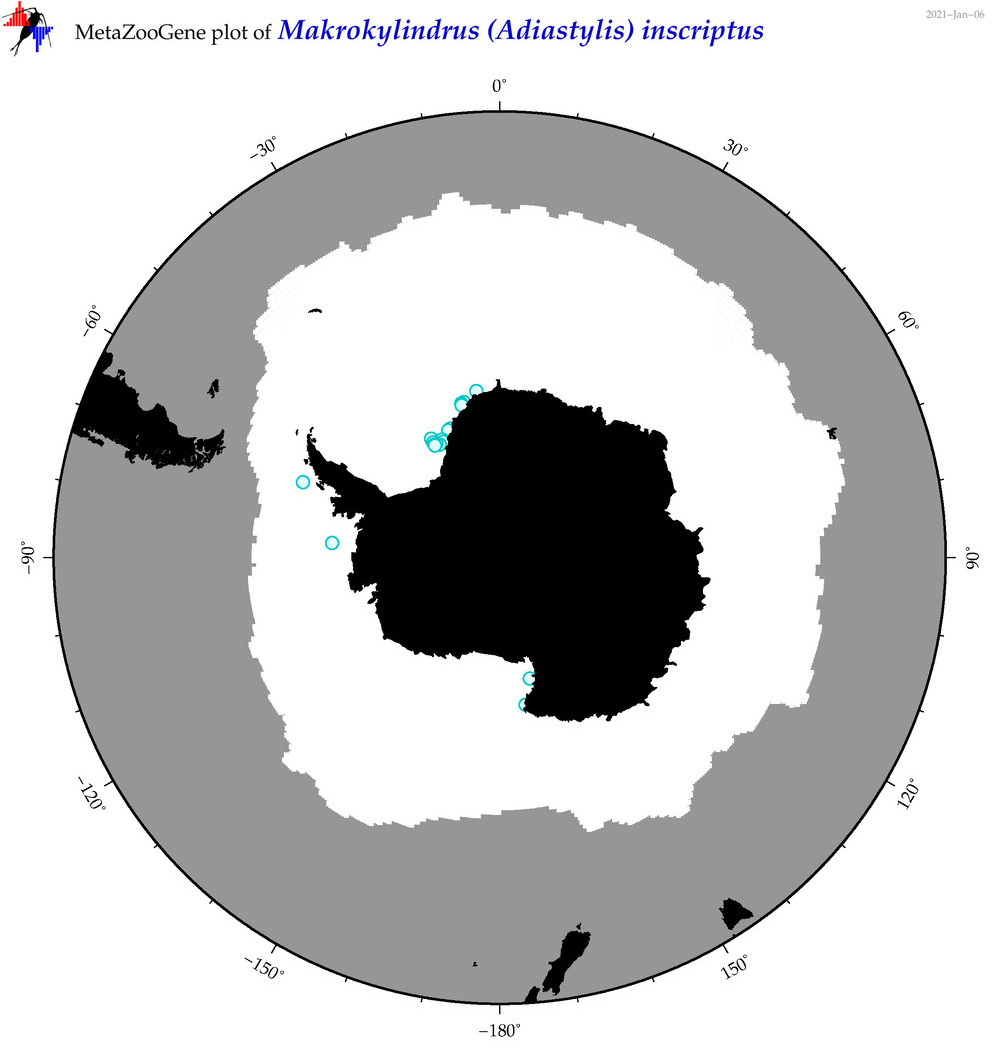 |
accepted
T4094817
R:1:1:0:0 |
| Paralamprops serratocostatus |
Species
(63) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4062257
R:1:0:0:0 |
| Platysympus brachyurus |
Species
(64) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4064257
R:1:0:0:0 |
| Platytyphlops aspera |
Species
(65) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
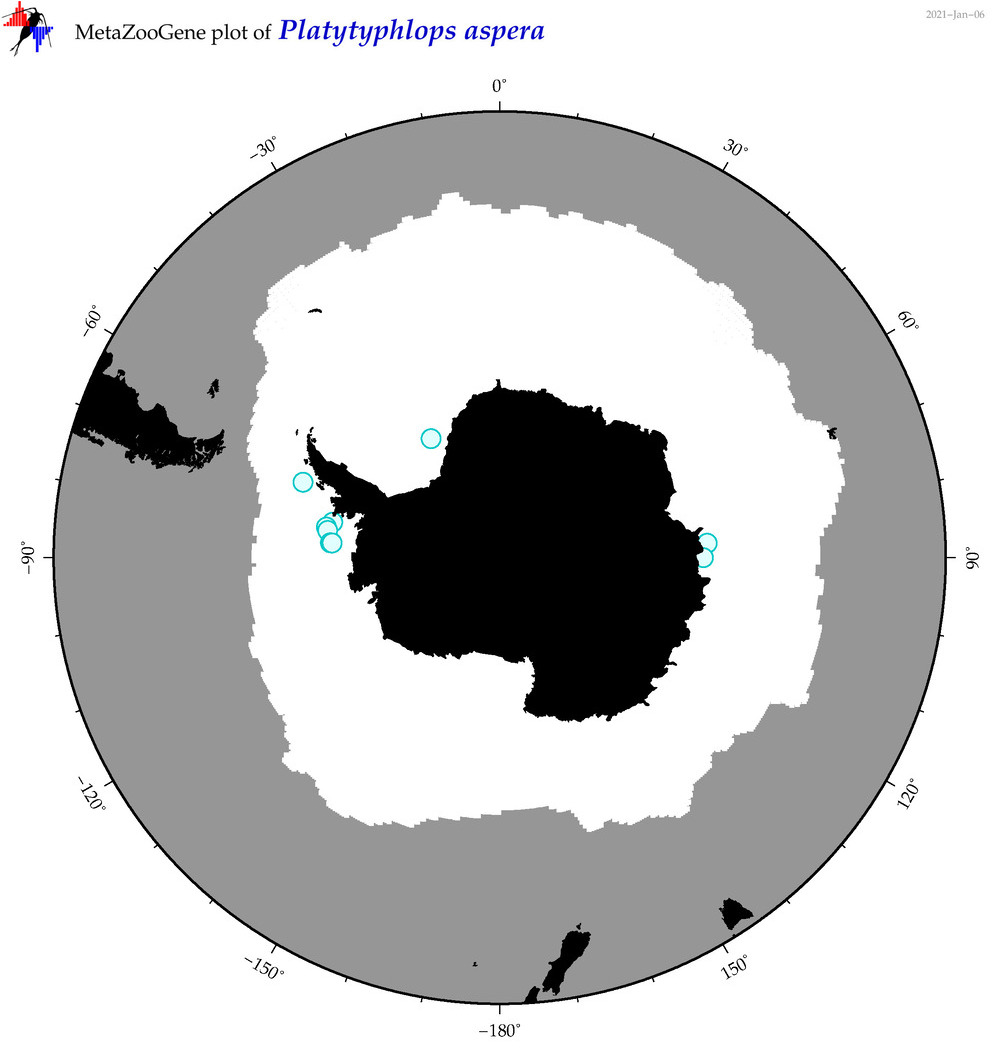 |
accepted
T4064271
R:1:0:0:0 |
| Platytyphlops mawsoni |
Species
(66) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
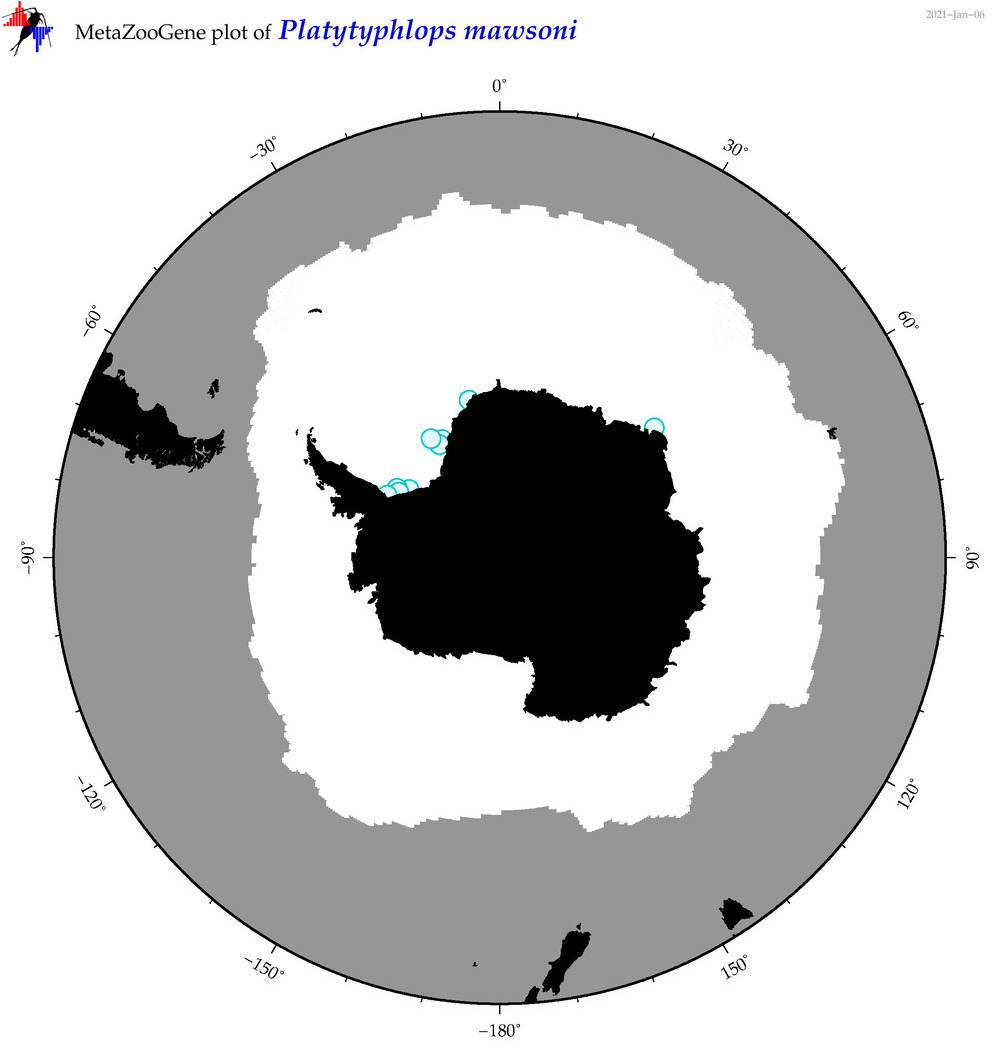 |
accepted
T4064279
R:1:0:0:0 |
| Platytyphlops racovitzai |
Species
(67) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
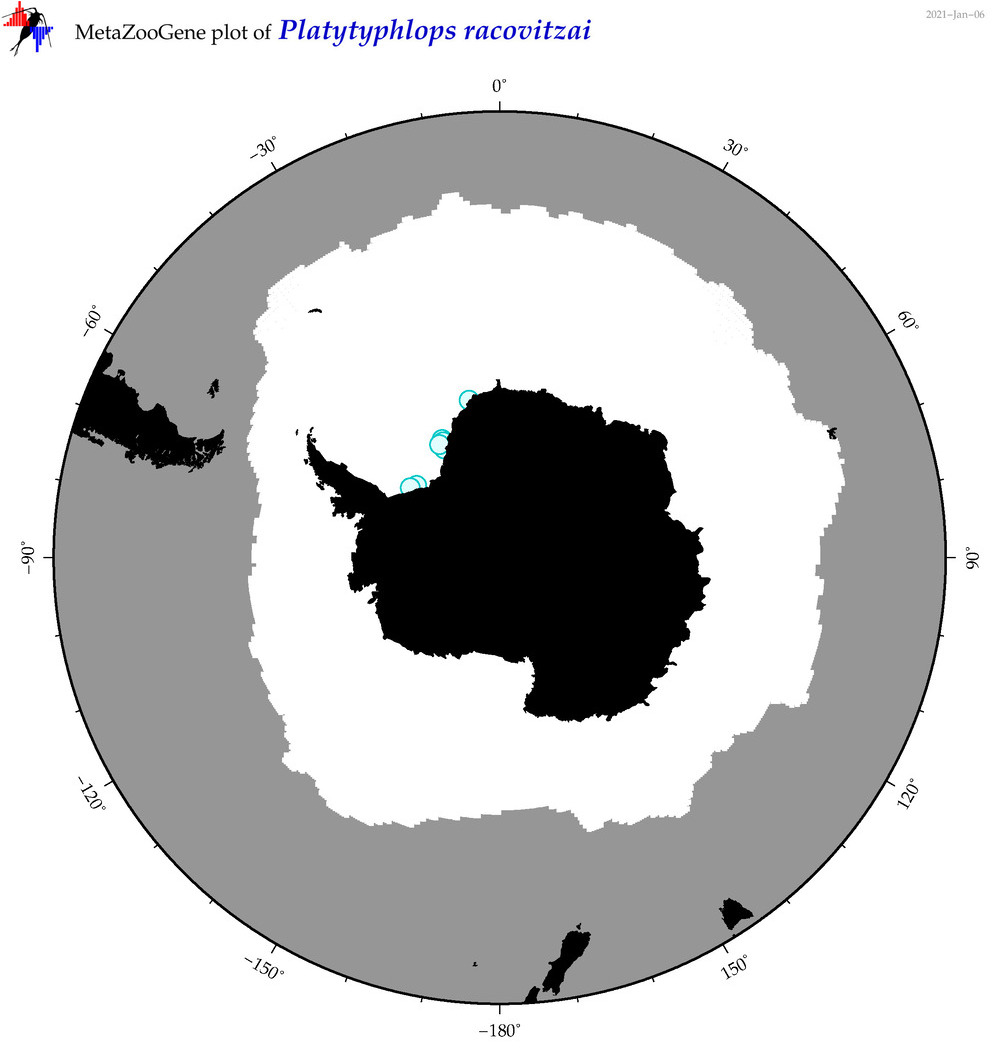 |
accepted
T4064282
R:1:0:0:0 |
| Platytyphlops rossi |
Species
(68) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
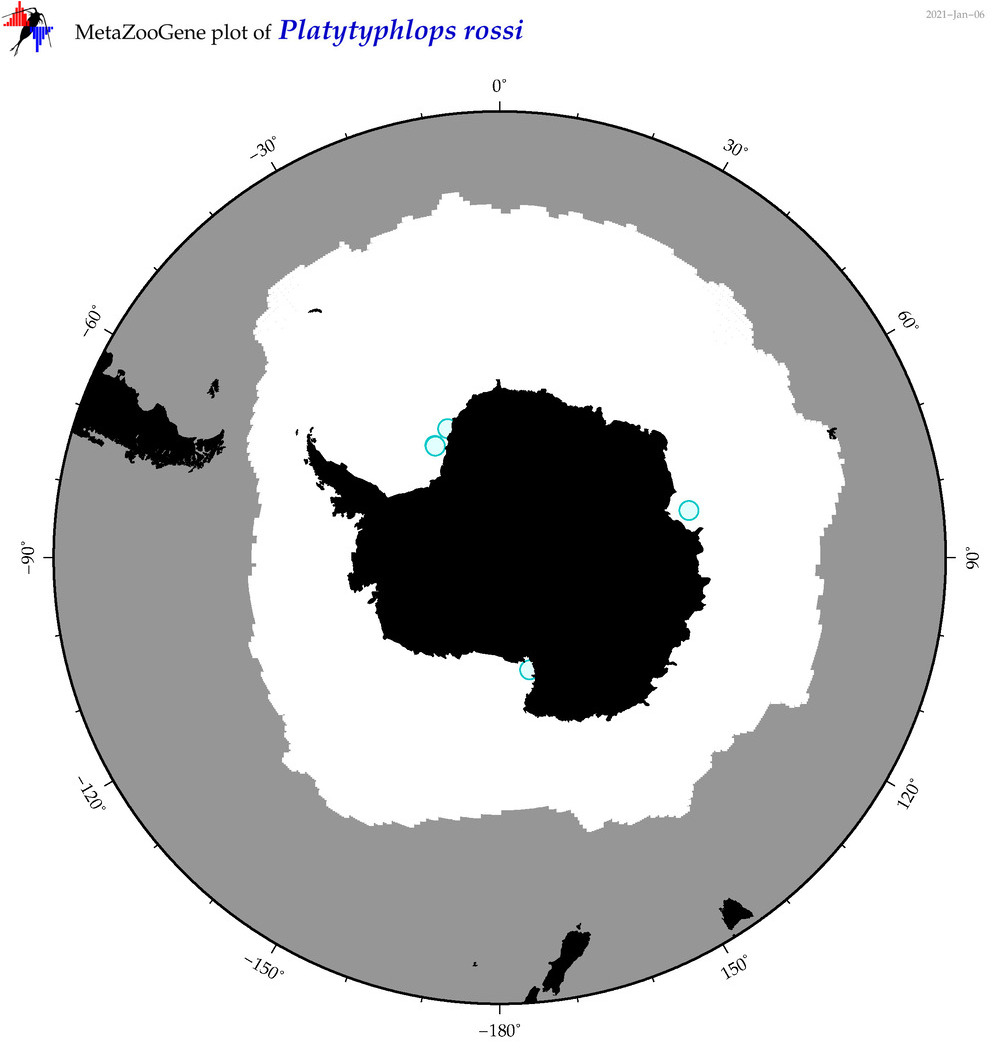 |
accepted
T4064283
R:1:0:0:0 |
| Procampylaspis armata |
Species
(69) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
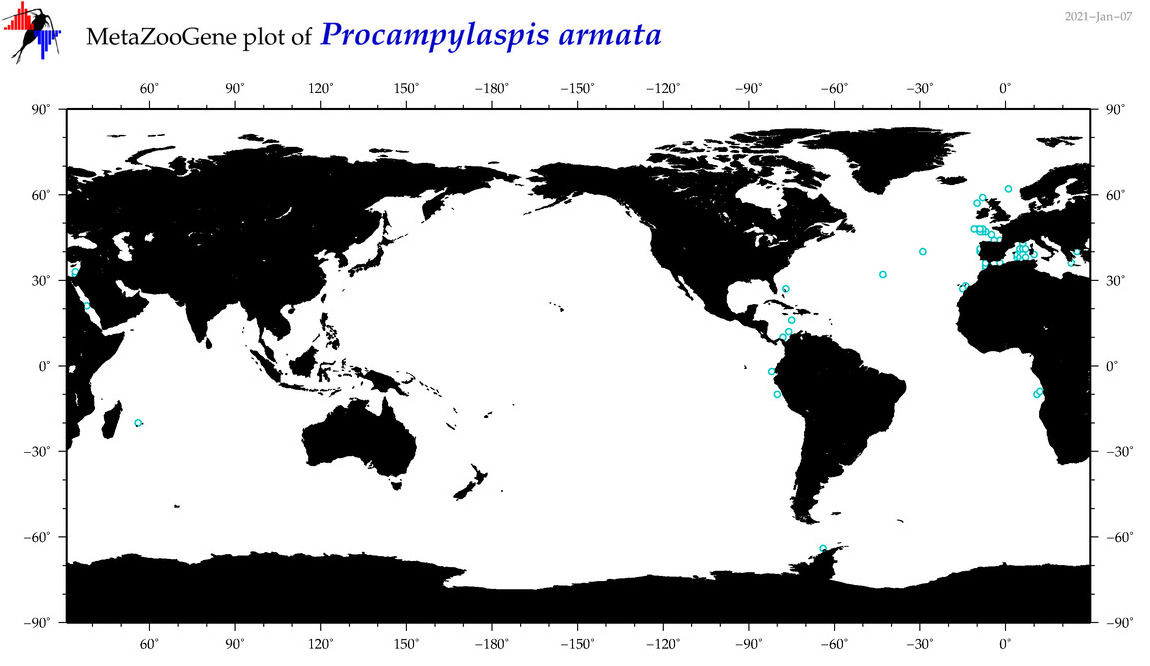 |
accepted
T4065355
R:1:0:0:0 |
| Procampylaspis compressa |
Species
(70) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
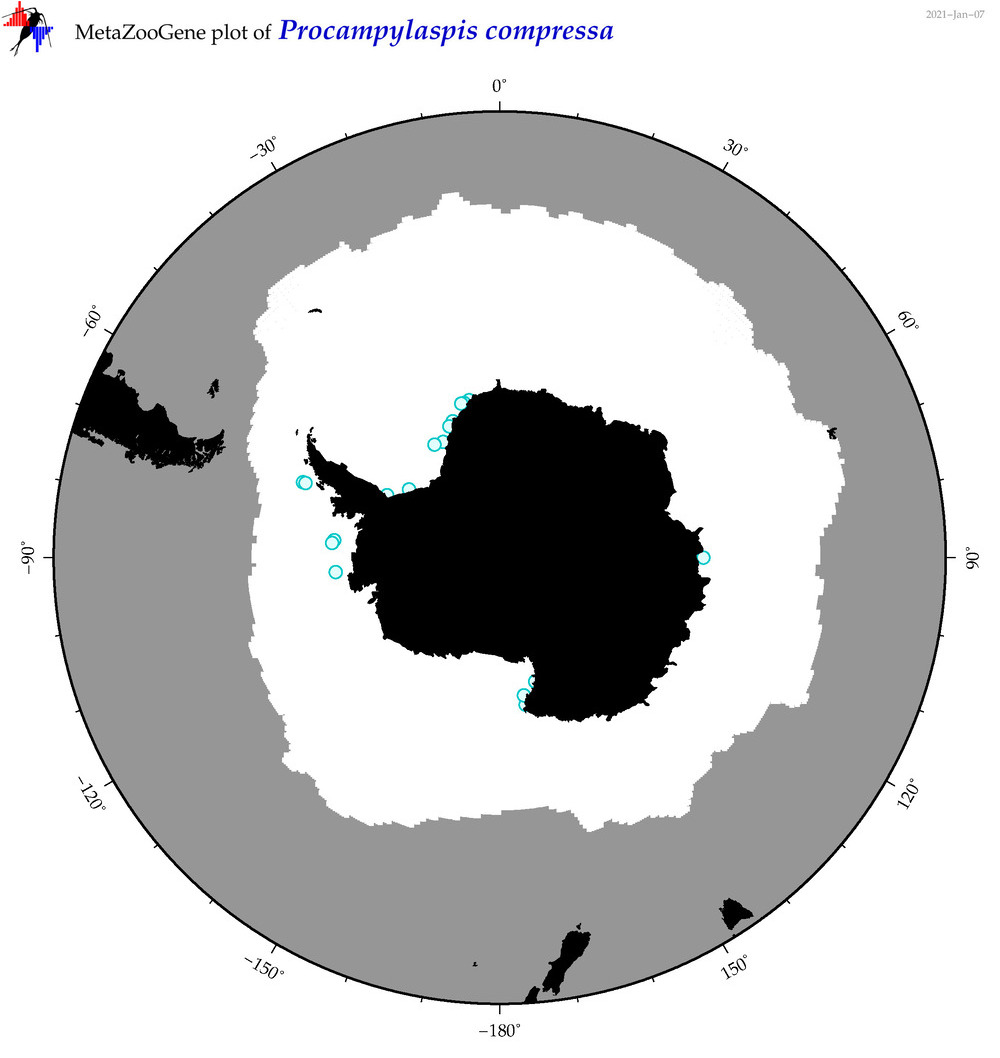 |
accepted
T4065364
R:1:0:0:0 |
| Procampylaspis halei |
Species
(71) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
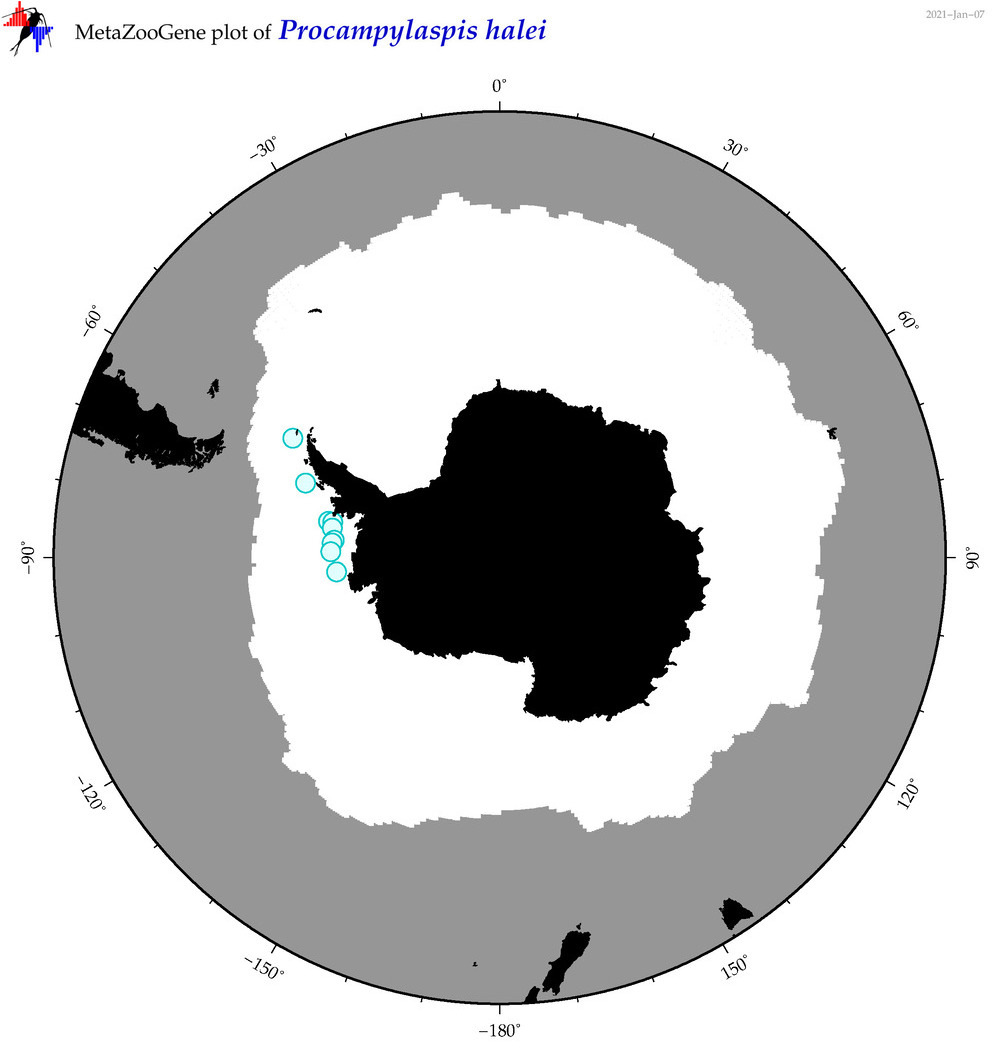 |
accepted
T4065368
R:1:0:0:0 |
| Procampylaspis poorei |
Species
(72) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
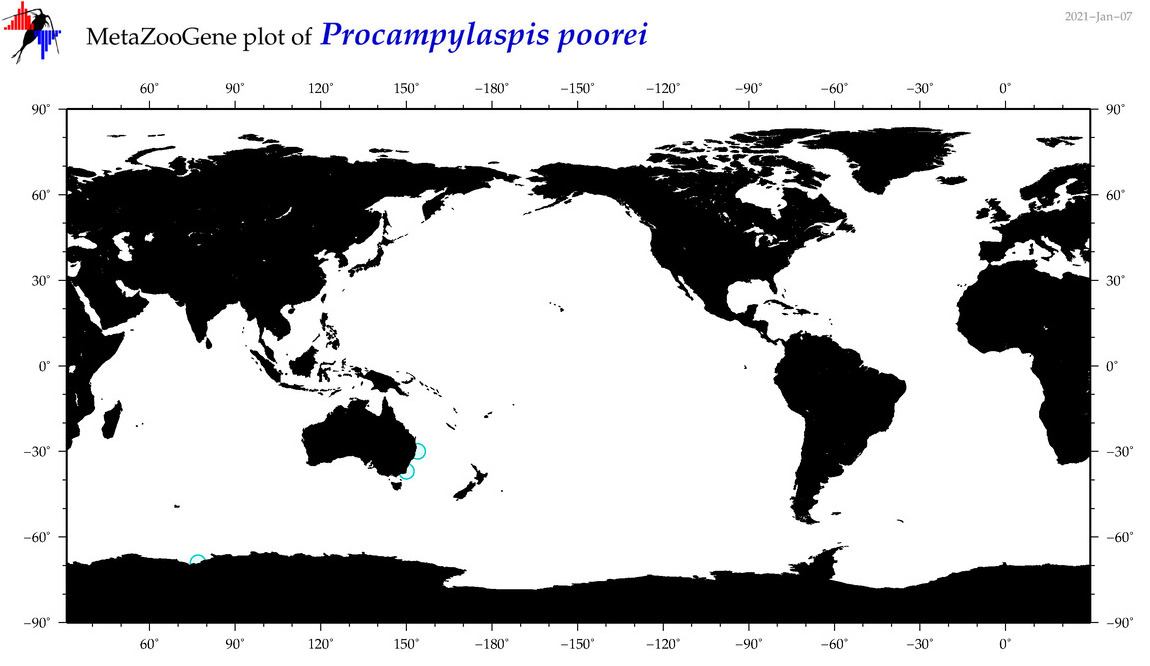 |
accepted
T4065377
R:1:0:0:0 |
| Schizocuma molossa |
Species
(73) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
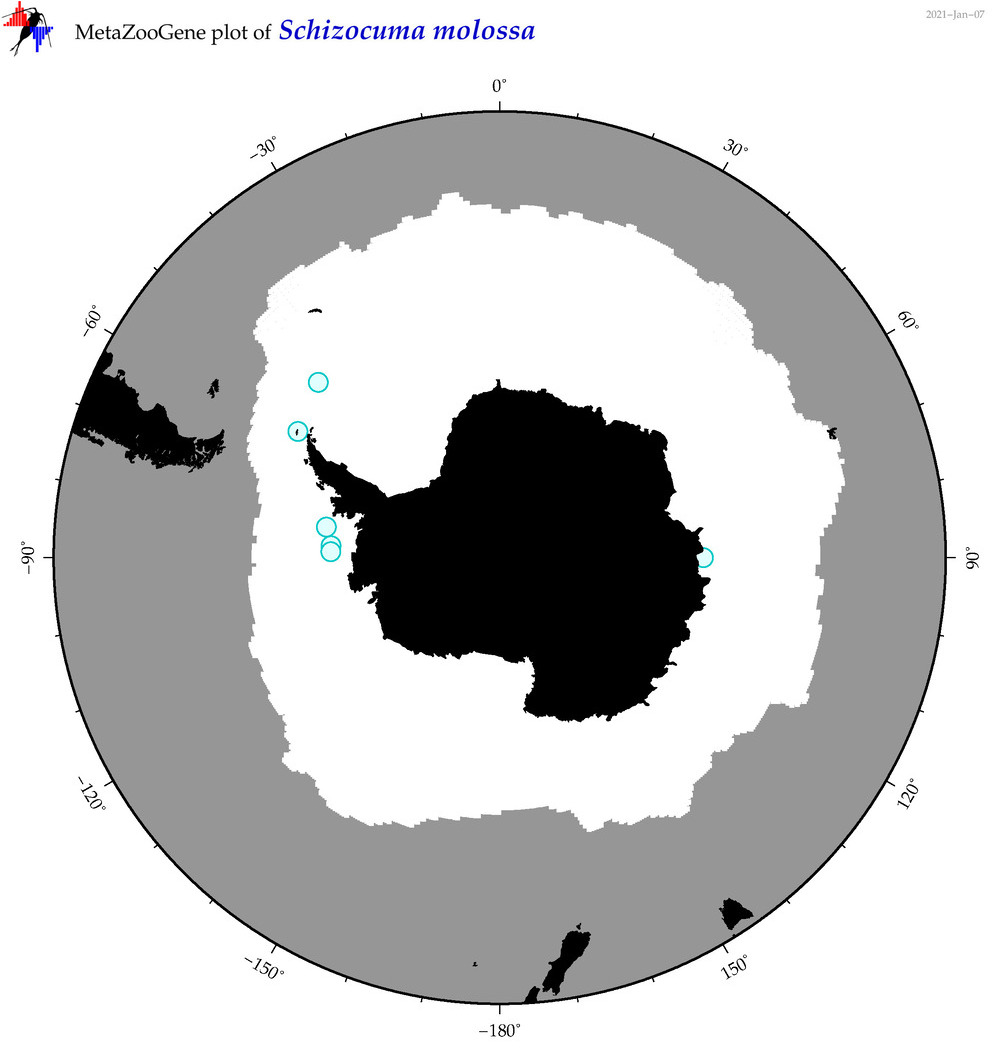 |
accepted
T4067381
R:1:0:0:0 |
| Vaunthompsonia inermis |
Species
(74) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
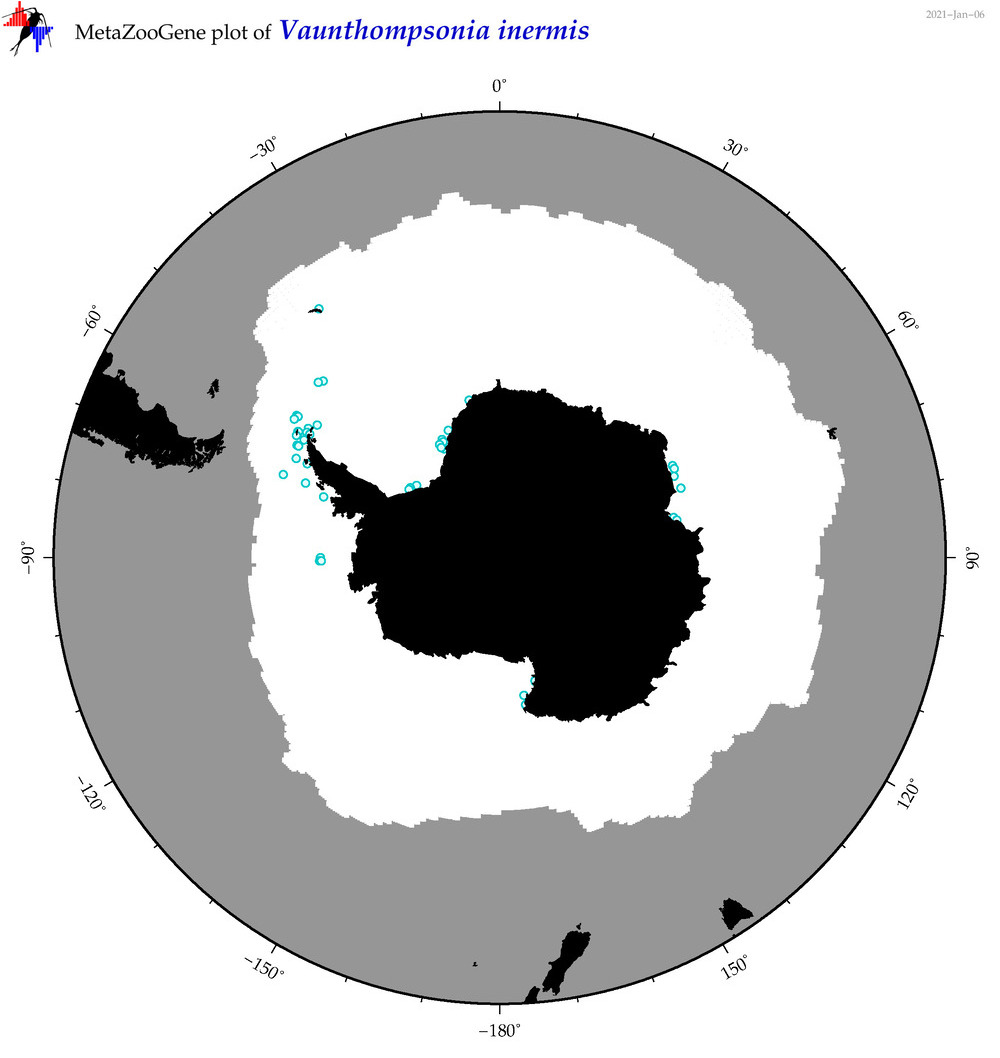 |
accepted
T4071385
R:1:0:0:0 |
| Vaunthompsonia laevifrons |
Species
(75) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
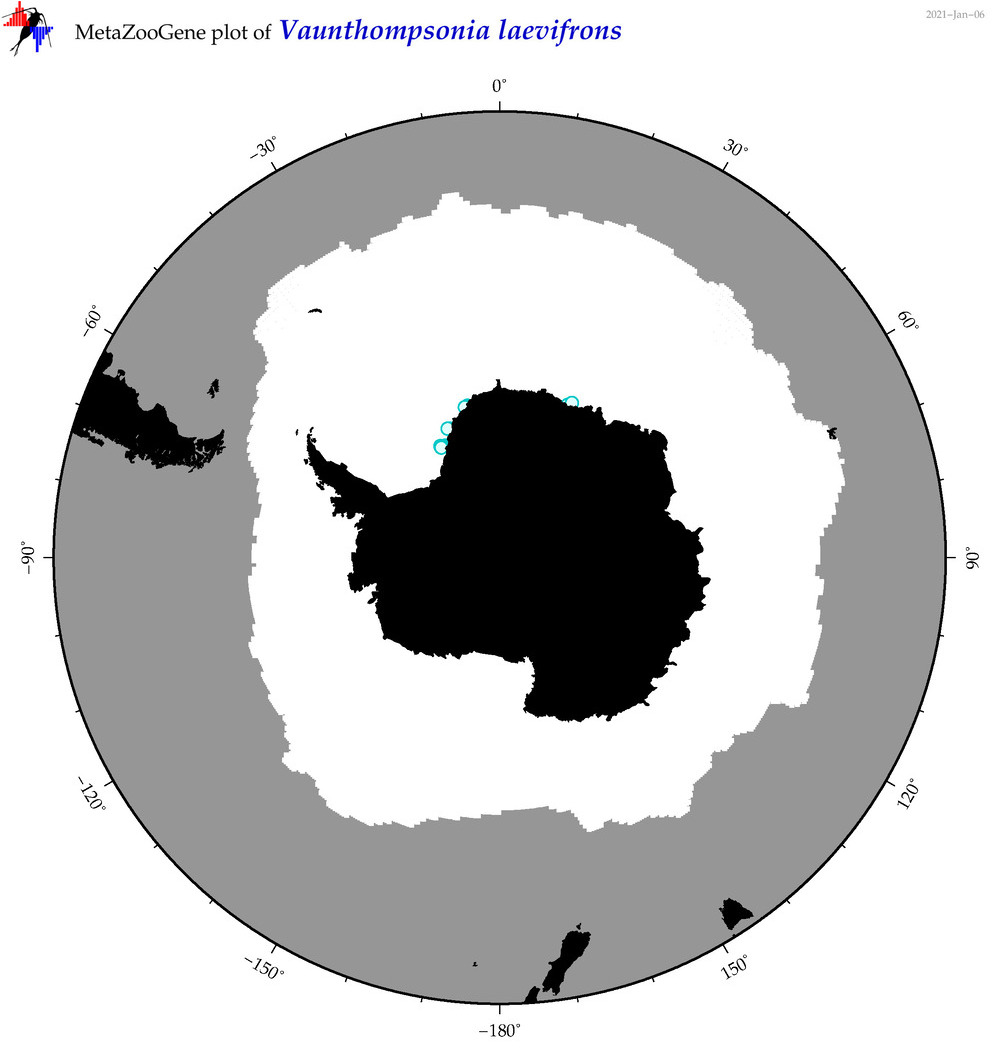 |
accepted
T4071386
R:1:0:0:0 |
| Vaunthompsonia meridionalis |
Species
(76) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4071387
R:1:0:0:0 |