Taxa Name
| Taxa
Level | Barcode
Data? | Number of
Barcodes
from
any ocean | Number of
Barcodes
from this region | Barcode Locations and
Species Occurence Map | MZGdb Atlas
v2026-m01-01
2026-Jan-24 |
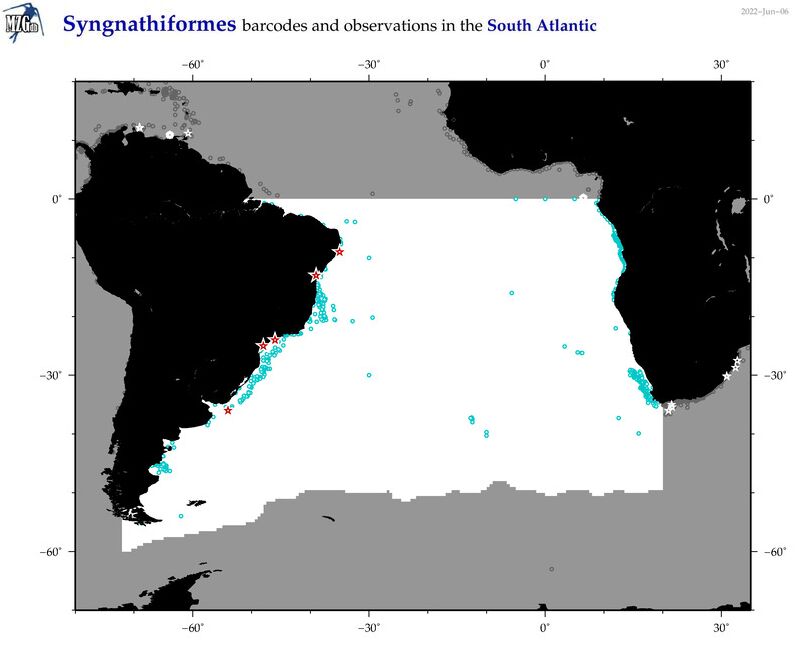
| Syngnathiformes |
Order |
cummulative
sub-totals
------->
for this
Order |
Total Species: 39
Sp. w/COI-Barcodes: 31
Total COI-Barcodes: 837 |
Total Species: 39
Sp. w/COI-Barcodes: 4
Total COI-Barcodes: 10 |
 |
accepted
T5002224
R:1:1:1:0 |
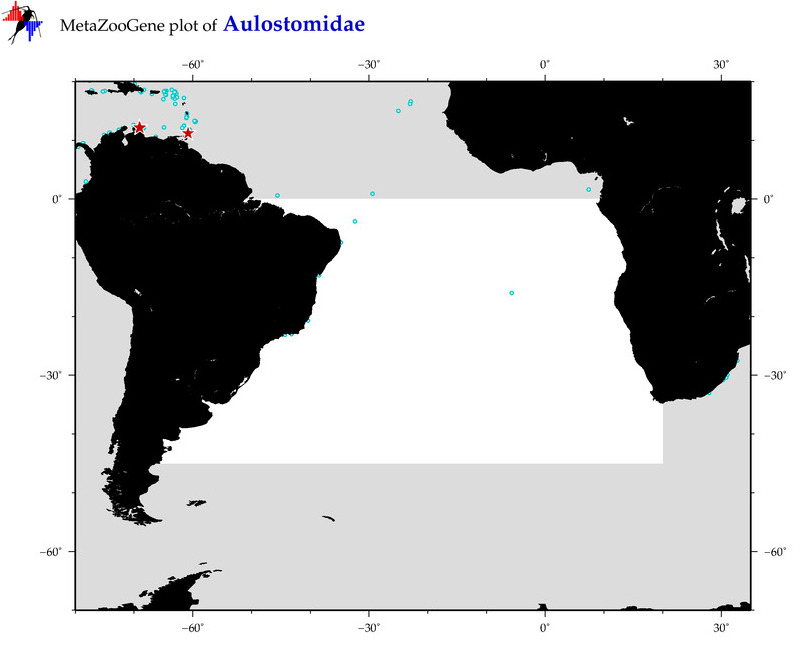
| ⋄ Aulostomidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 14 |
Total Species: 2
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5000237
R:1:0:0:0 |
| ⋄ ⋄ Aulostomus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 14 |
Total Species: 2
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5002165
R:1:0:0:0 |
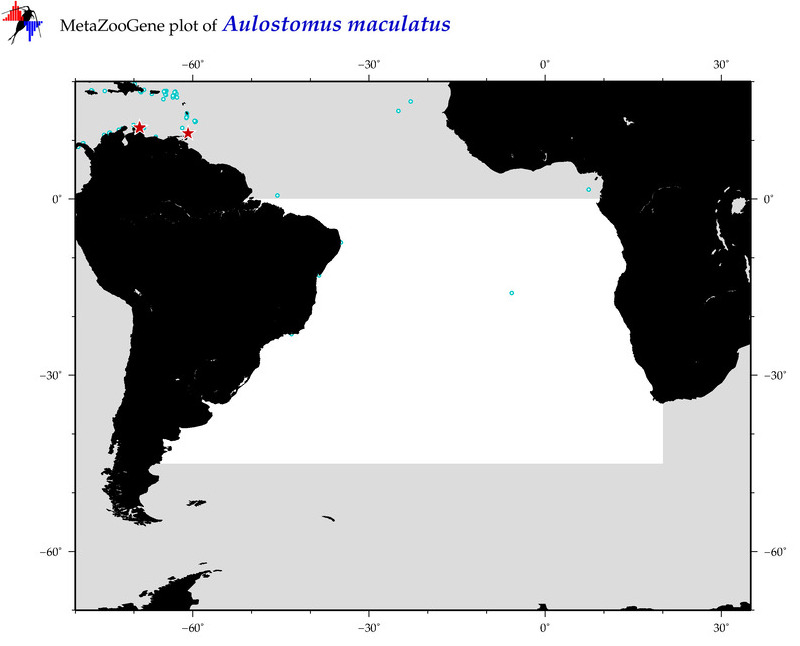
| ⋄ ⋄ ⋄ Aulostomus maculatus |
Species
(1) |
COI
12S
16S
18S
28S
|
COI = 14
12S = 2
16S = 4
18S = 1
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5002166
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Aulostomus strigosus |
Species
(2) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5016848
R:1:0:0:0 |
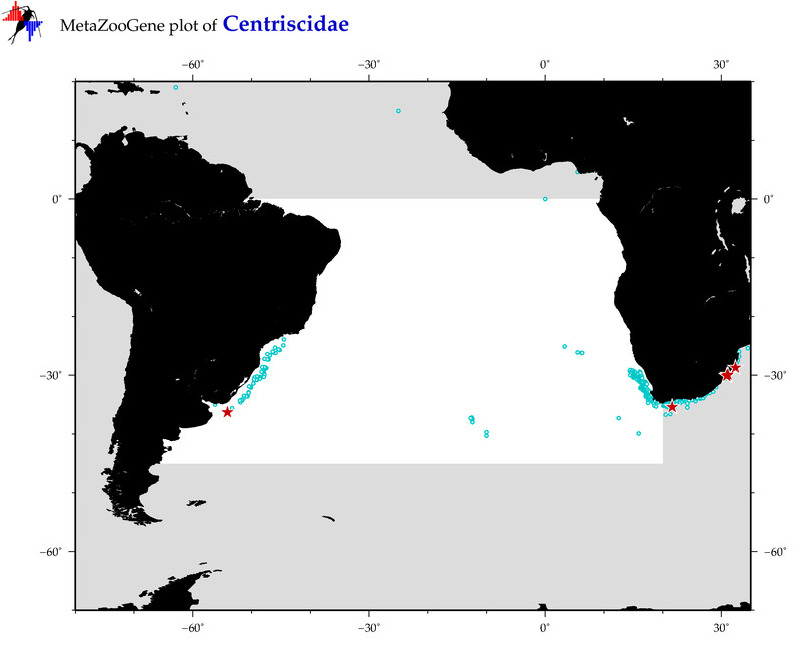
| ⋄ Centriscidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 7
Sp. w/COI-Barcodes: 5
Total COI-Barcodes: 108 |
Total Species: 7
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 2 |
 |
accepted
T5000355
R:1:1:0:0 |
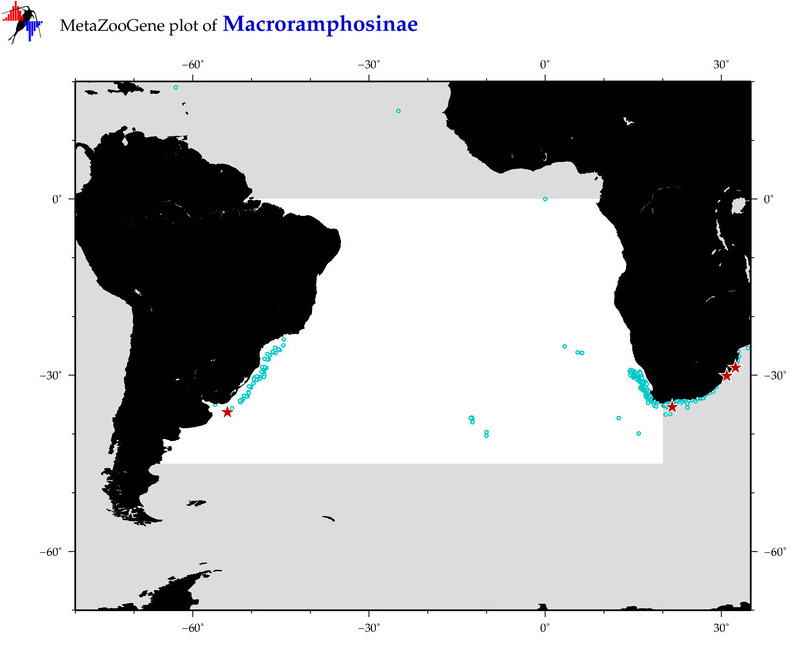
| ⋄ ⋄ Macroramphosinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 7
Sp. w/COI-Barcodes: 5
Total COI-Barcodes: 108 |
Total Species: 7
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 2 |
 |
accepted
T5002355
R:1:0:0:0 |
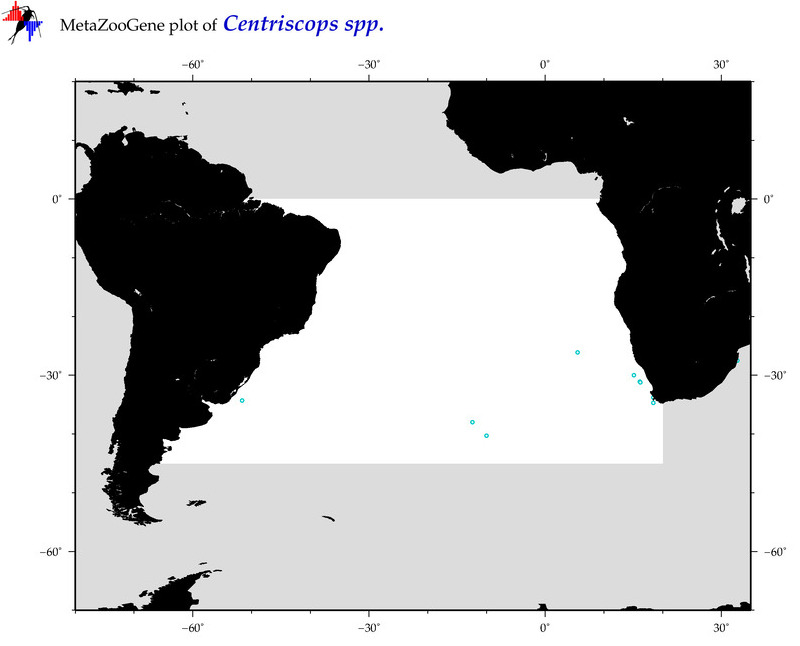
| ⋄ ⋄ ⋄ Centriscops |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5002508
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Centriscops humerosus |
Species
(3) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003425
R:1:0:0:0 |
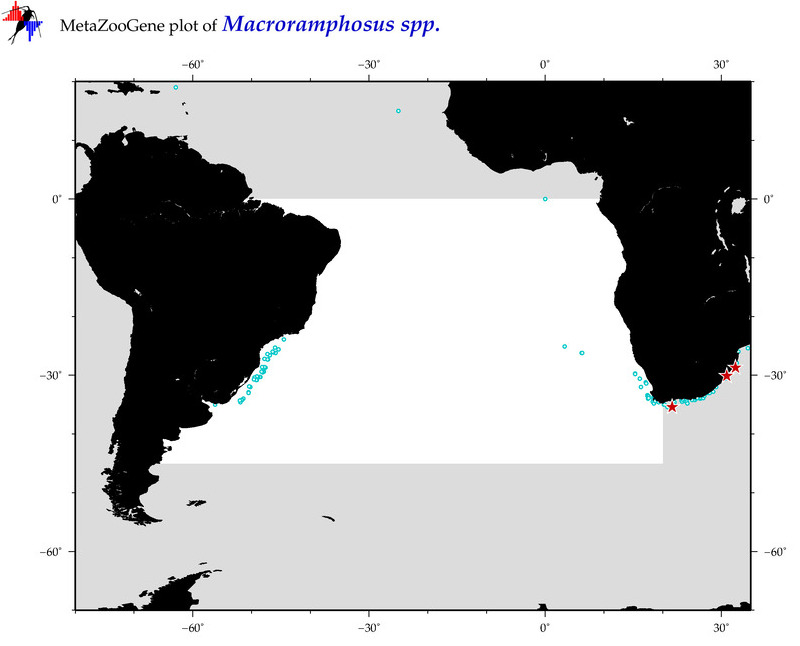
| ⋄ ⋄ ⋄ Macroramphosus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 104 |
Total Species: 2
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5000857
R:1:0:0:0 |
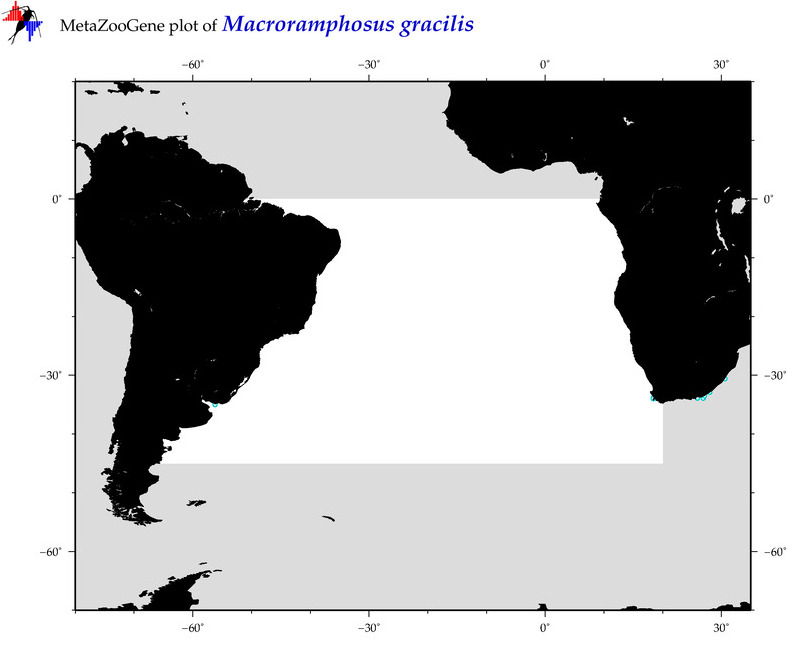
| ⋄ ⋄ ⋄ ⋄ Macroramphosus gracilis |
Species
(4) |
COI
12S
16S
18S
28S
|
COI = 17
12S = 5
16S = 4
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000856
R:1:0:0:0 |
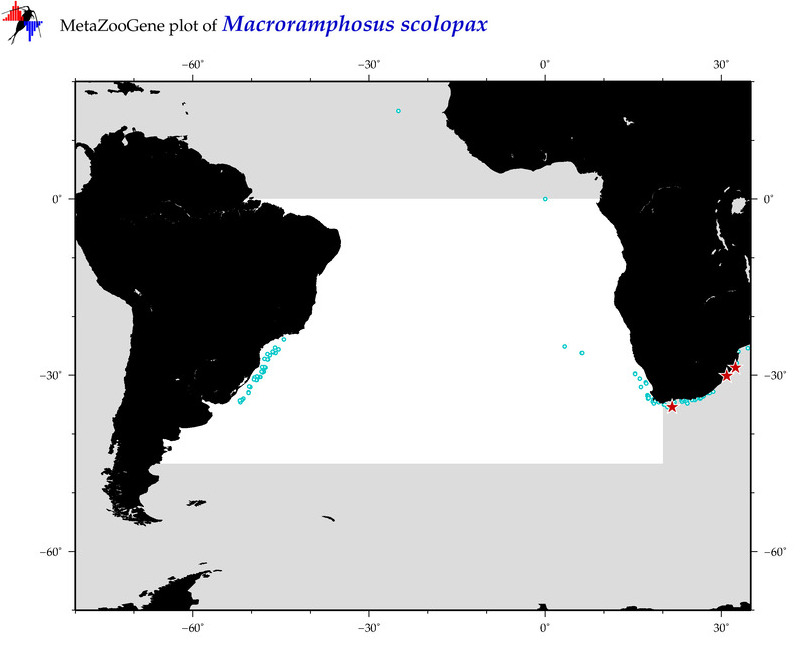
| ⋄ ⋄ ⋄ ⋄ Macroramphosus scolopax |
Species
(5) |
COI
12S
16S
18S
28S
|
COI = 87
12S = 6
16S = 20
18S = 0
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000858
R:1:0:0:0 |
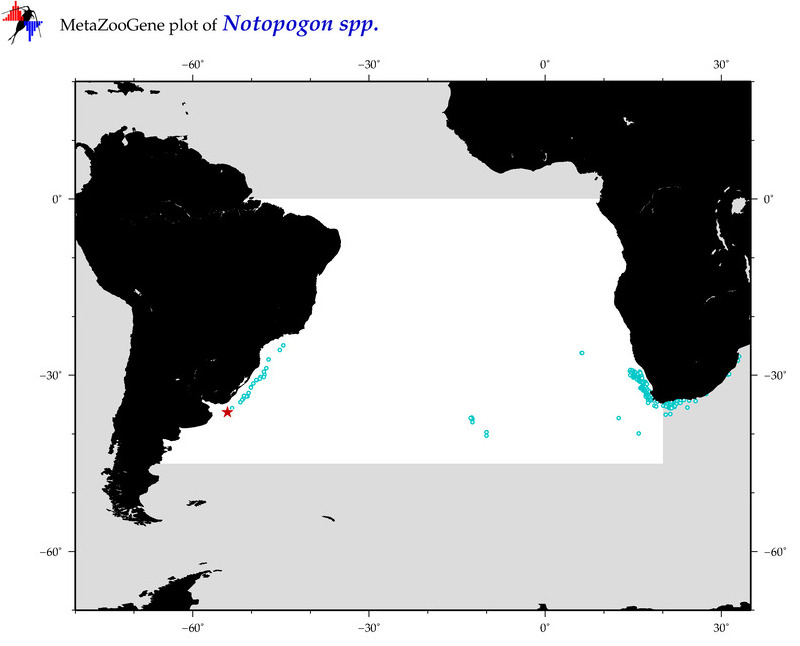
| ⋄ ⋄ ⋄ Notopogon |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 4
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 3 |
Total Species: 4
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 2 |
 |
accepted
T5002798
R:1:0:0:0 |
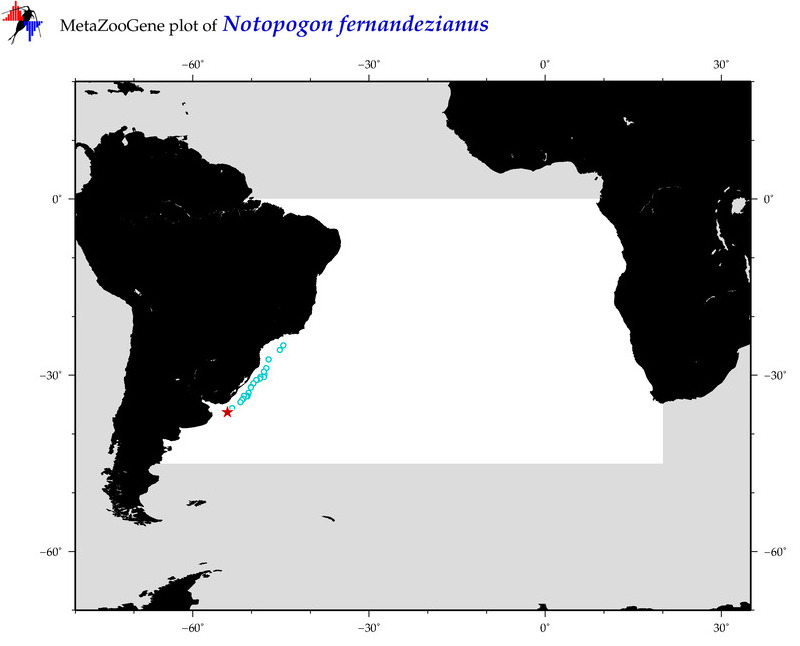
| ⋄ ⋄ ⋄ ⋄ Notopogon fernandezianus |
Species
(6) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004814
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Notopogon lilliei |
Species
(7) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004815
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Notopogon macrosolen |
Species
(8) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022789
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Notopogon xenosoma |
Species
(9) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022790
R:1:0:0:0 |
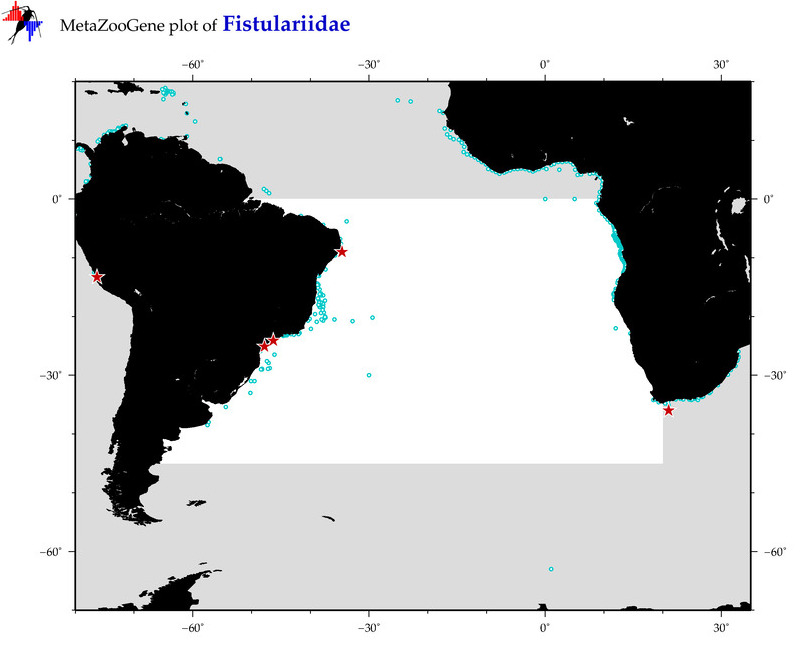
| ⋄ Fistulariidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 55 |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 7 |
 |
accepted
T5000584
R:1:1:0:0 |
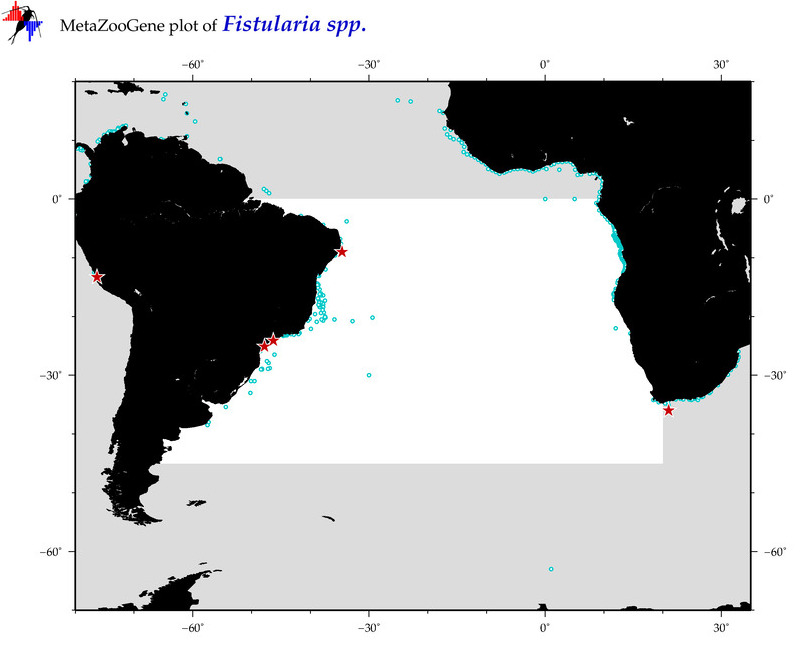
| ⋄ ⋄ Fistularia |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 55 |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 7 |
 |
accepted
T5000583
R:1:1:0:0 |
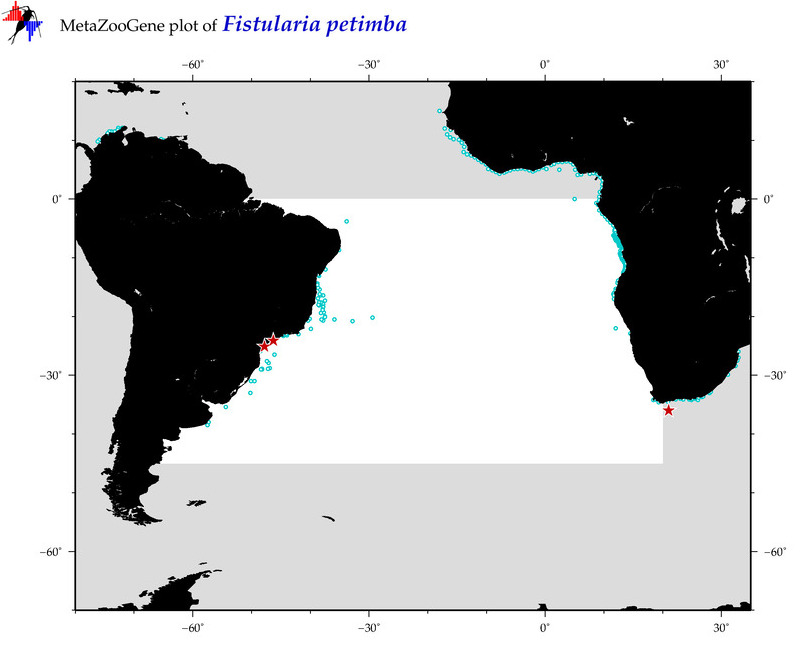
| ⋄ ⋄ ⋄ Fistularia petimba |
Species
(10) |
COI
12S
16S
18S
28S
|
COI = 45
12S = 15
16S = 11
18S = 1
28S = 0
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003998
R:1:1:0:0 |
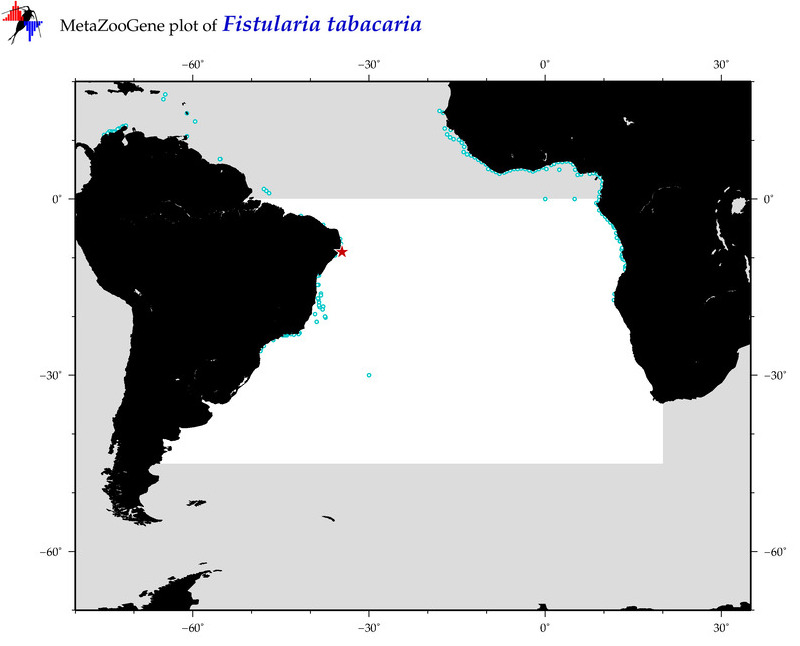
| ⋄ ⋄ ⋄ Fistularia tabacaria |
Species
(11) |
COI
12S
16S
18S
28S
|
COI = 10
12S = 2
16S = 2
18S = 0
28S = 0
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003999
R:1:1:0:0 |
| ⋄ Solenostomidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 9 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5005992
R:1:0:0:0 |
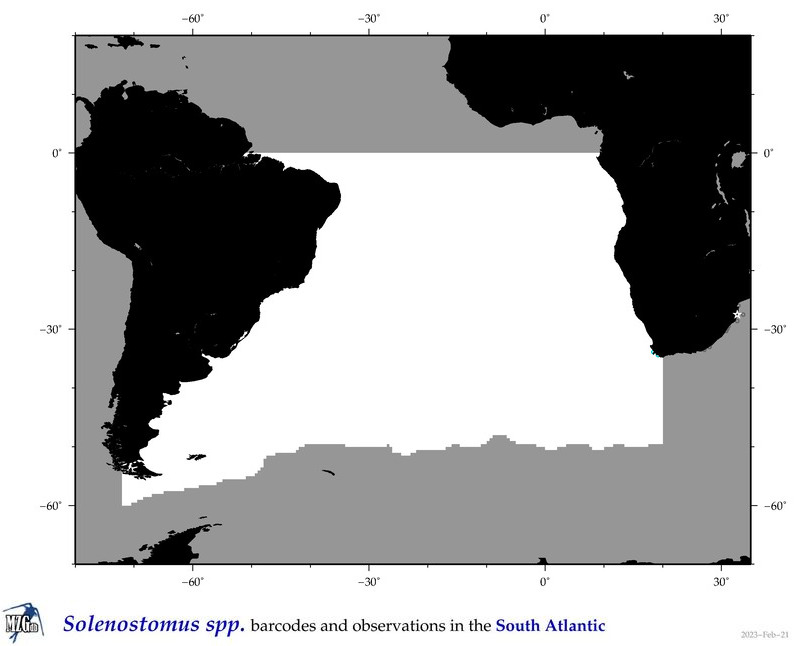
| ⋄ ⋄ Solenostomus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 9 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5006359
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Solenostomus cyanopterus |
Species
(12) |
COI
12S
16S
18S
28S
|
COI = 9
12S = 5
16S = 3
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5012272
R:1:0:0:0 |
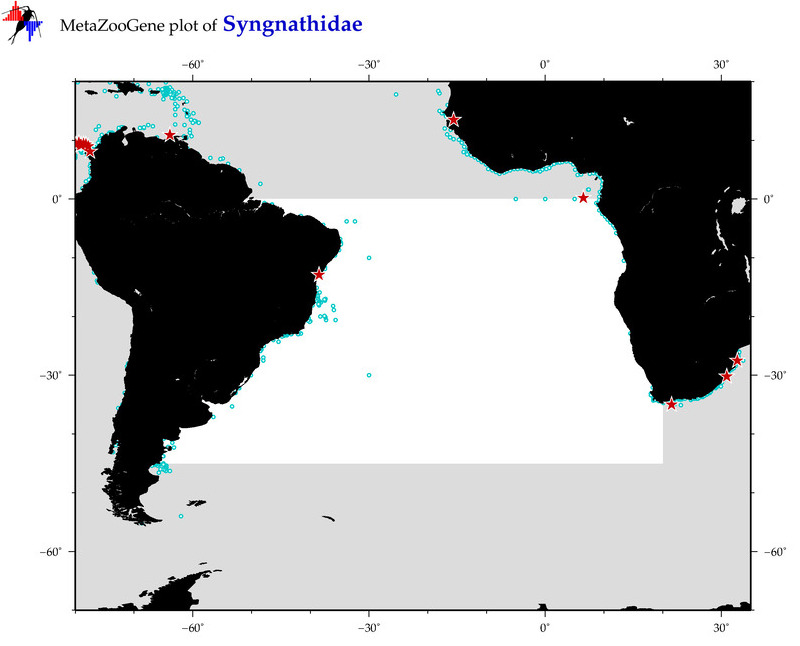
| ⋄ Syngnathidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 27
Sp. w/COI-Barcodes: 22
Total COI-Barcodes: 651 |
Total Species: 27
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
 |
accepted
T5000121
R:1:1:1:0 |
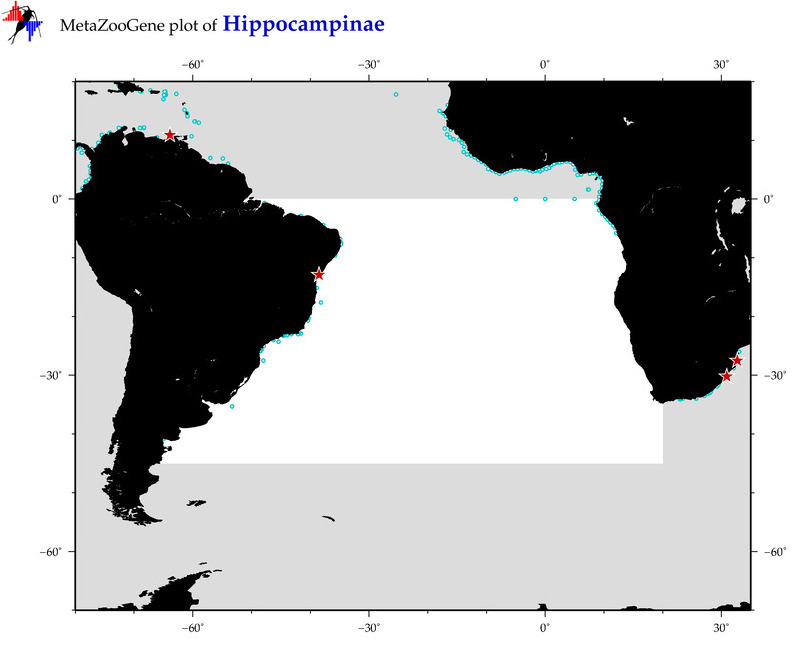
| ⋄ ⋄ Hippocampinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 7
Sp. w/COI-Barcodes: 7
Total COI-Barcodes: 383 |
Total Species: 7
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
 |
accepted
T5002357
R:1:1:0:0 |
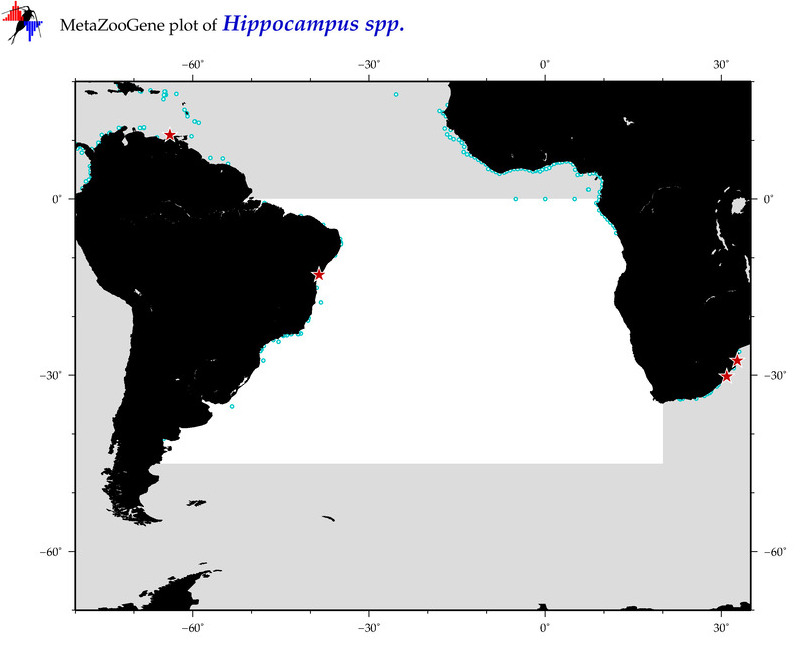
| ⋄ ⋄ ⋄ Hippocampus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 7
Sp. w/COI-Barcodes: 7
Total COI-Barcodes: 383 |
Total Species: 7
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
 |
accepted
T5000668
R:1:1:0:0 |
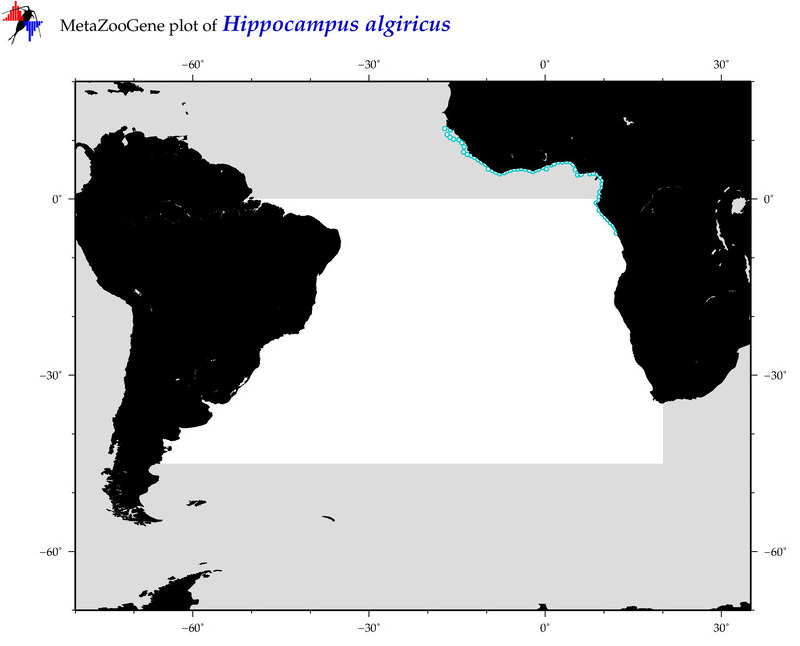
| ⋄ ⋄ ⋄ ⋄ Hippocampus algiricus |
Species
(13) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 2
16S = 4
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004251
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hippocampus barbouri |
Species
(14) |
COI
12S
16S
18S
28S
|
COI = 76
12S = 2
16S = 10
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
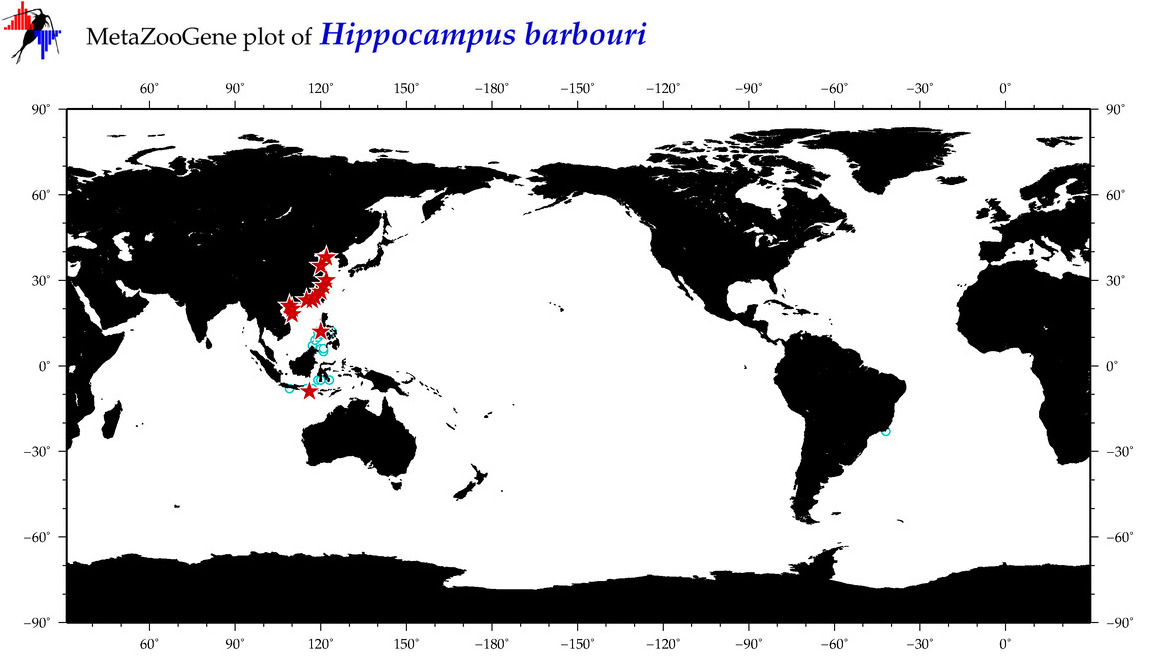 |
accepted
T5004253
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hippocampus erectus |
Species
(15) |
COI
12S
16S
18S
28S
|
COI = 49
12S = 5
16S = 18
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
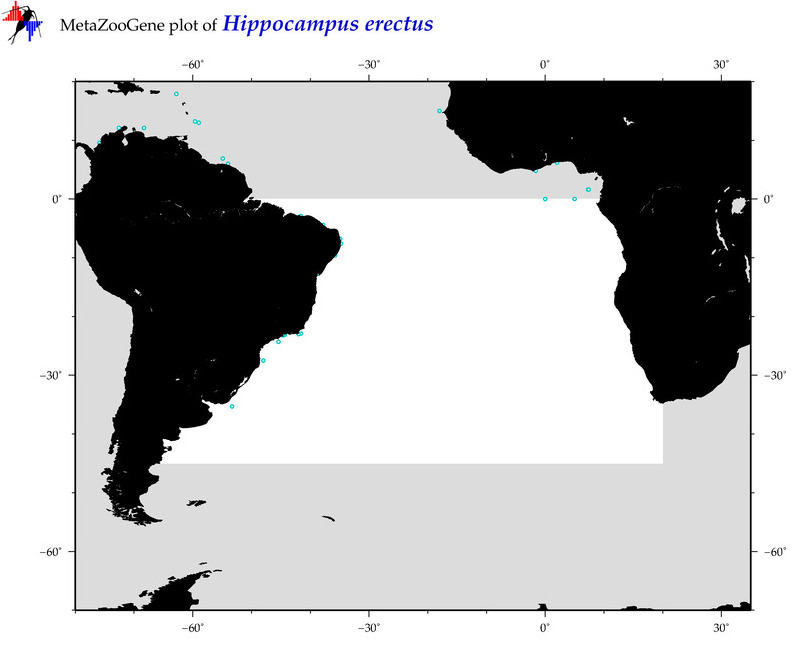 |
accepted
T5000669
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hippocampus guttulatus |
Species
(16) |
COI
12S
16S
18S
28S
|
COI = 12
12S = 1
16S = 8
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
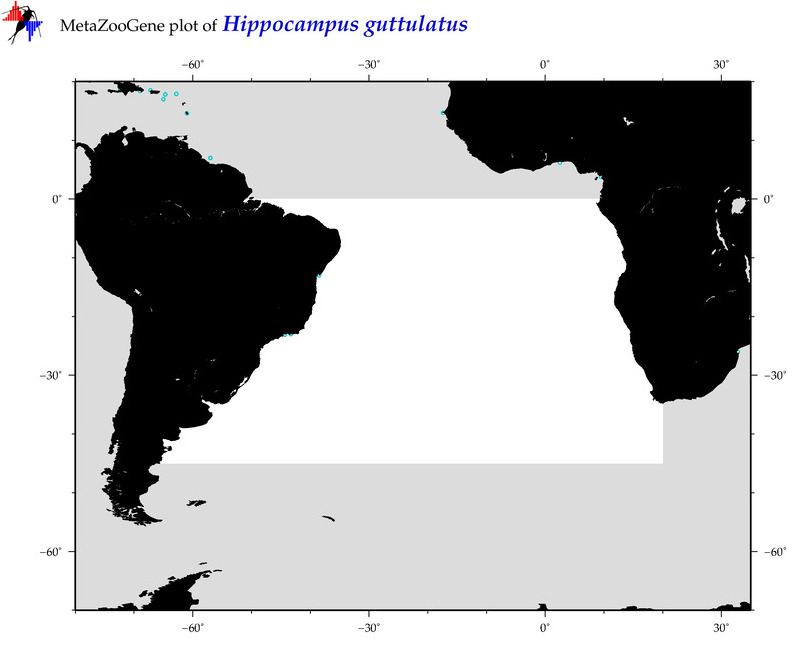 |
accepted
T5004262
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hippocampus hippocampus |
Species
(17) |
COI
12S
16S
18S
28S
|
COI = 48
12S = 3
16S = 24
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004264
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hippocampus kuda |
Species
(18) |
COI
12S
16S
18S
28S
|
COI = 140
12S = 10
16S = 45
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
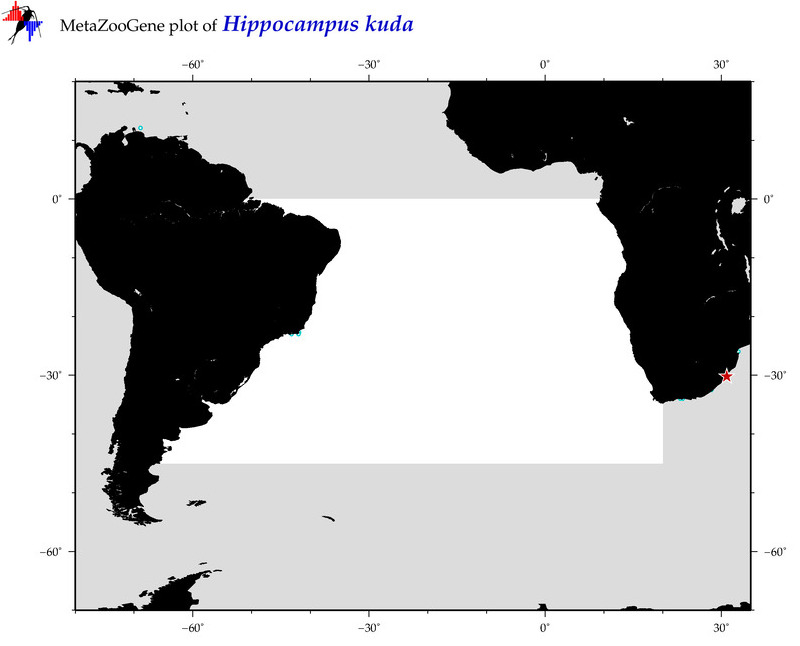 |
accepted
T5004269
R:1:1:0:0 |
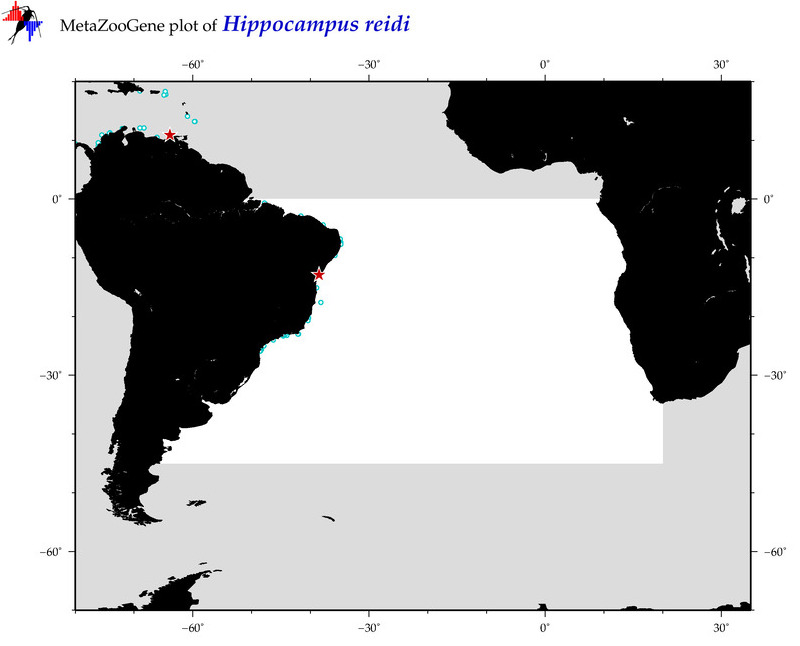
| ⋄ ⋄ ⋄ ⋄ Hippocampus reidi |
Species
(19) |
COI
12S
16S
18S
28S
|
COI = 54
12S = 4
16S = 9
18S = 1
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004274
R:1:1:0:0 |
| ⋄ ⋄ Syngnathinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 20
Sp. w/COI-Barcodes: 15
Total COI-Barcodes: 268 |
Total Species: 20
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
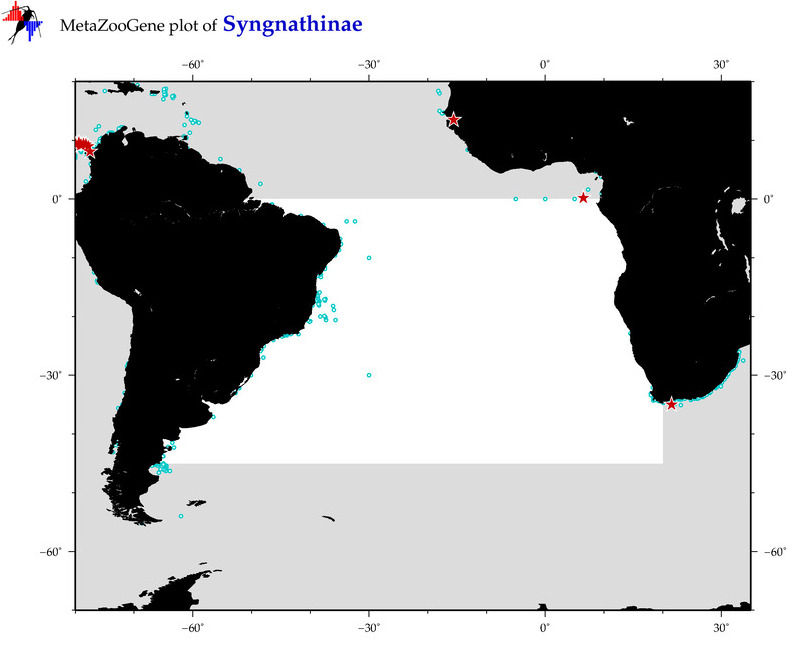 |
accepted
T5002356
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Amphelikturus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 3 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
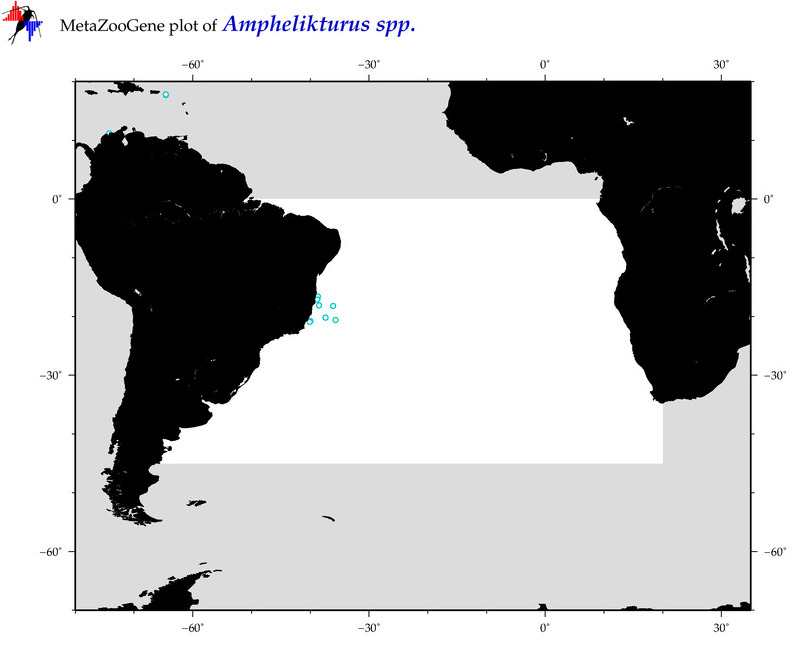 |
accepted
T5002440
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Amphelikturus dendriticus |
Species
(20) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 1
16S = 1
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
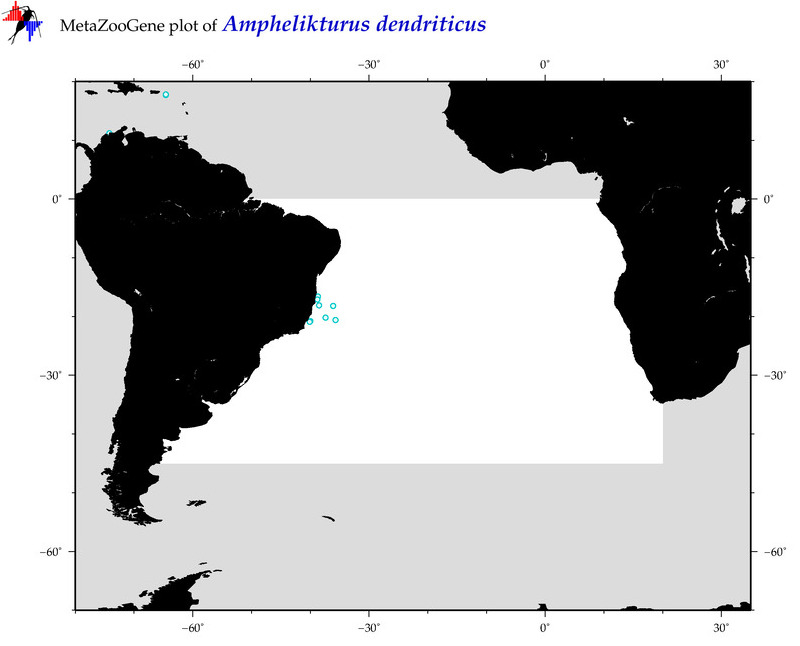 |
accepted
T5003086
R:1:0:0:0 |
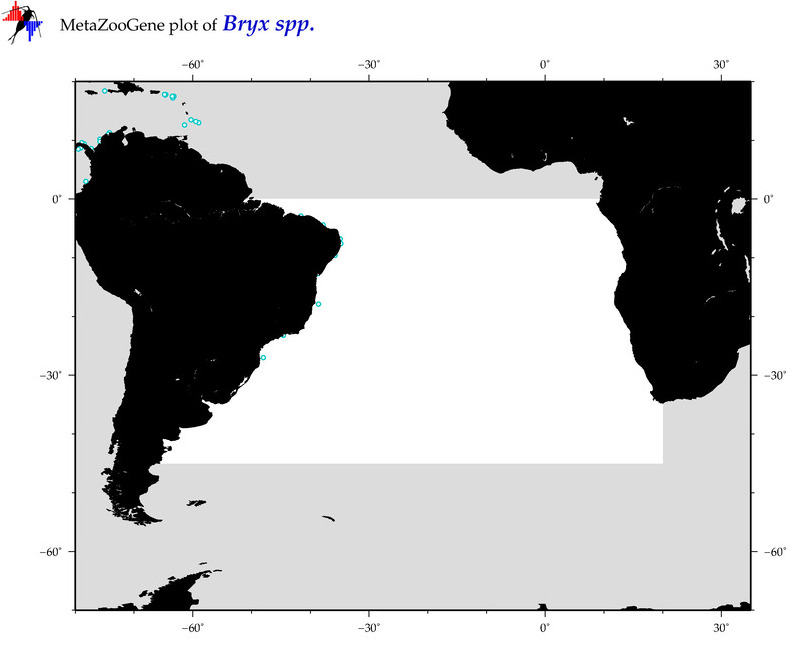
| ⋄ ⋄ ⋄ Bryx |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 20 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5002498
R:1:0:0:0 |
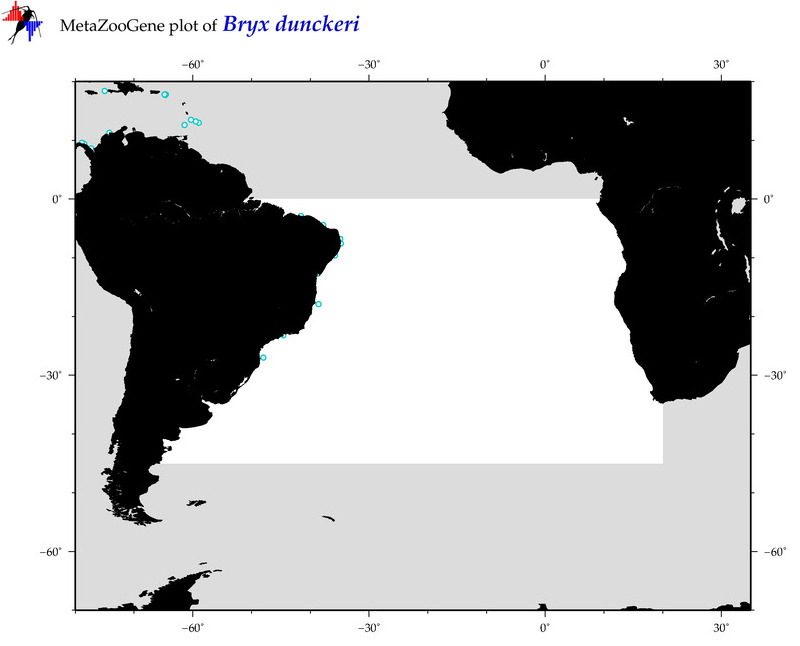
| ⋄ ⋄ ⋄ ⋄ Bryx dunckeri |
Species
(21) |
COI
12S
16S
18S
28S
|
COI = 20
12S = 2
16S = 2
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003319
R:1:0:0:0 |
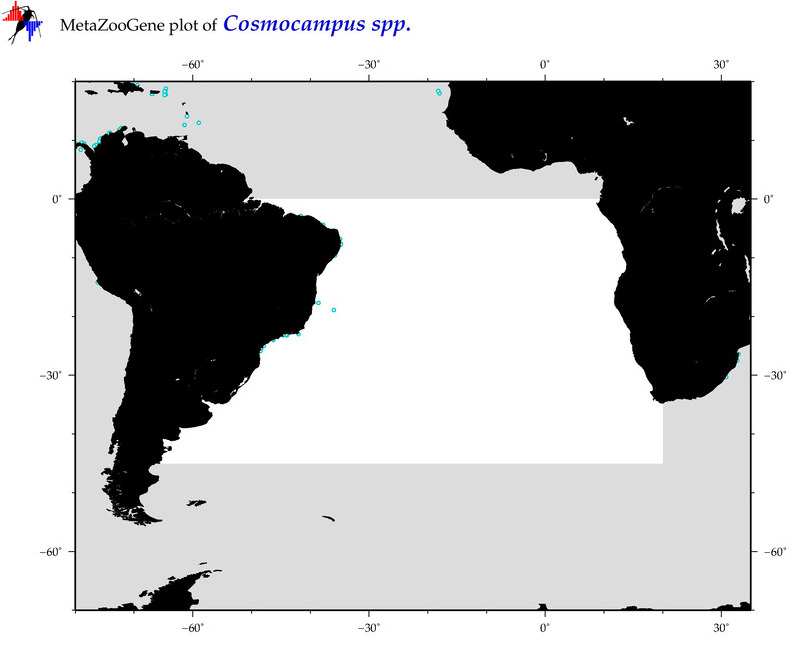
| ⋄ ⋄ ⋄ Cosmocampus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 3
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 17 |
Total Species: 3
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5002546
R:1:0:0:0 |
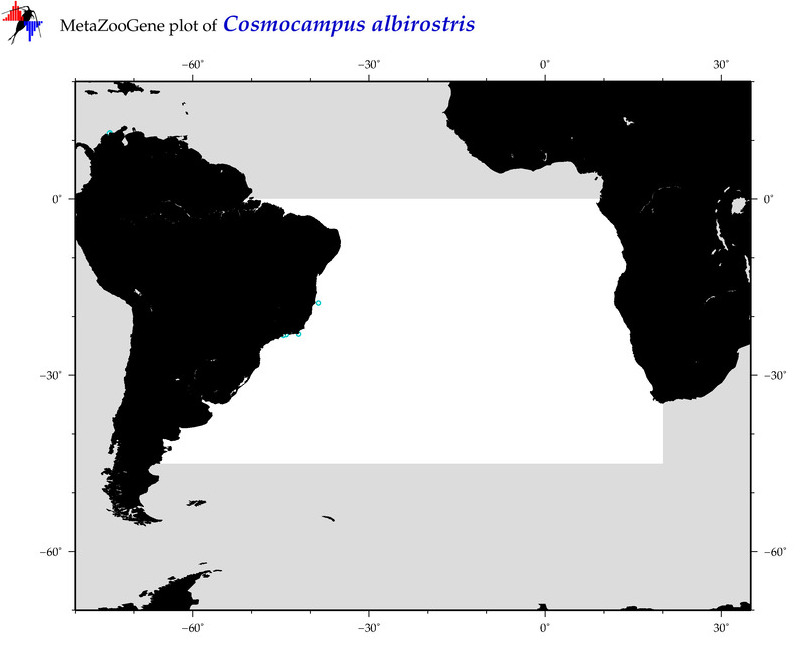
| ⋄ ⋄ ⋄ ⋄ Cosmocampus albirostris |
Species
(22) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 1
16S = 1
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003649
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Cosmocampus elucens |
Species
(23) |
COI
12S
16S
18S
28S
|
COI = 13
12S = 3
16S = 3
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003653
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Cosmocampus profundus |
Species
(24) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5018683
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Enneacampus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5002602
R:0:1:1:0 |
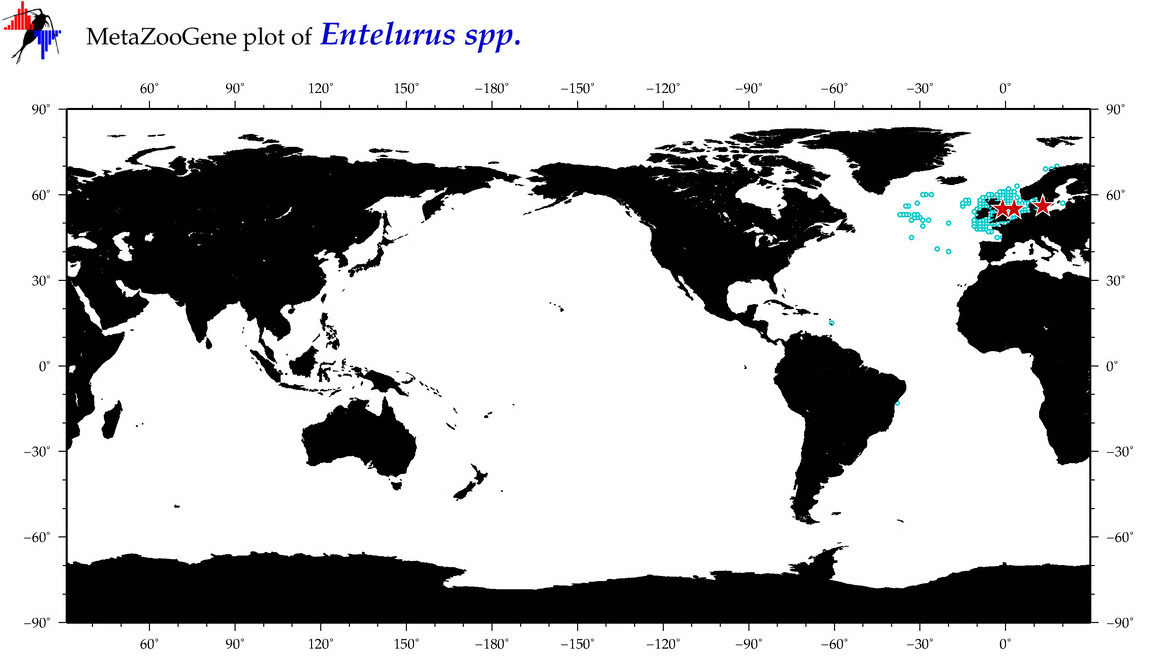
| ⋄ ⋄ ⋄ Entelurus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 10 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5002603
R:1:1:0:0 |
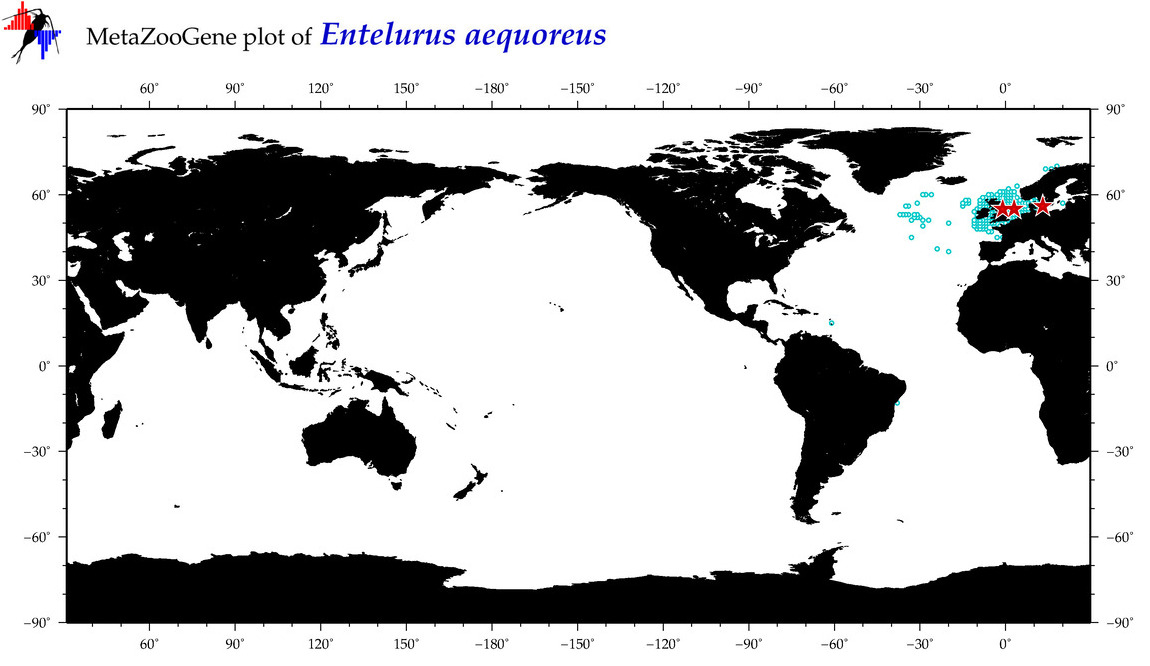
| ⋄ ⋄ ⋄ ⋄ Entelurus aequoreus |
Species
(25) |
COI
12S
16S
18S
28S
|
COI = 10
12S = 3
16S = 5
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003877
R:1:1:0:0 |
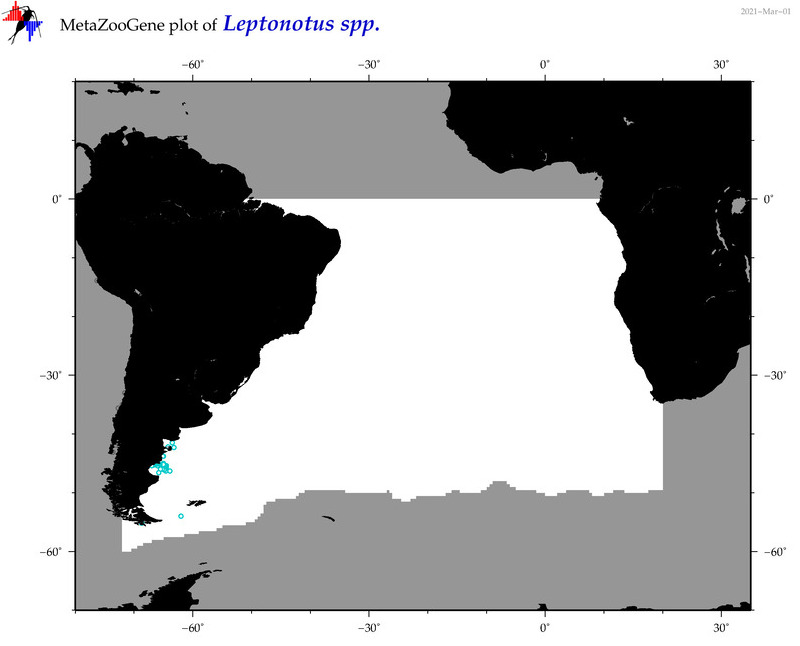
| ⋄ ⋄ ⋄ Leptonotus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5012844
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Leptonotus blainvilleanus |
Species
(26) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5021546
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Micrognathus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
Total Species: 2
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5002762
R:1:0:0:0 |
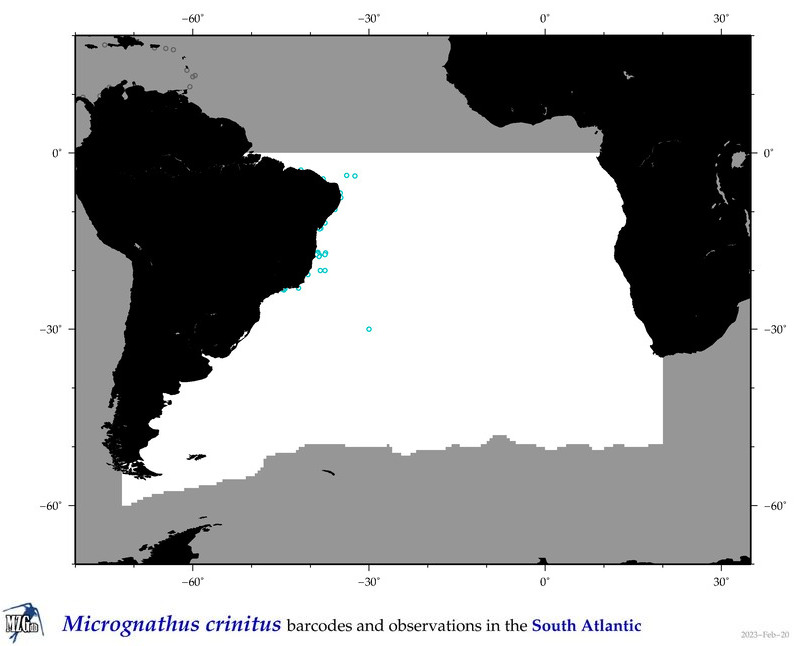
| ⋄ ⋄ ⋄ ⋄ Micrognathus crinitus |
Species
(27) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 1
16S = 1
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5022182
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Micrognathus erugatus |
Species
(28) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022183
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Microphis |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 3
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 115 |
Total Species: 3
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5002765
R:1:1:1:0 |
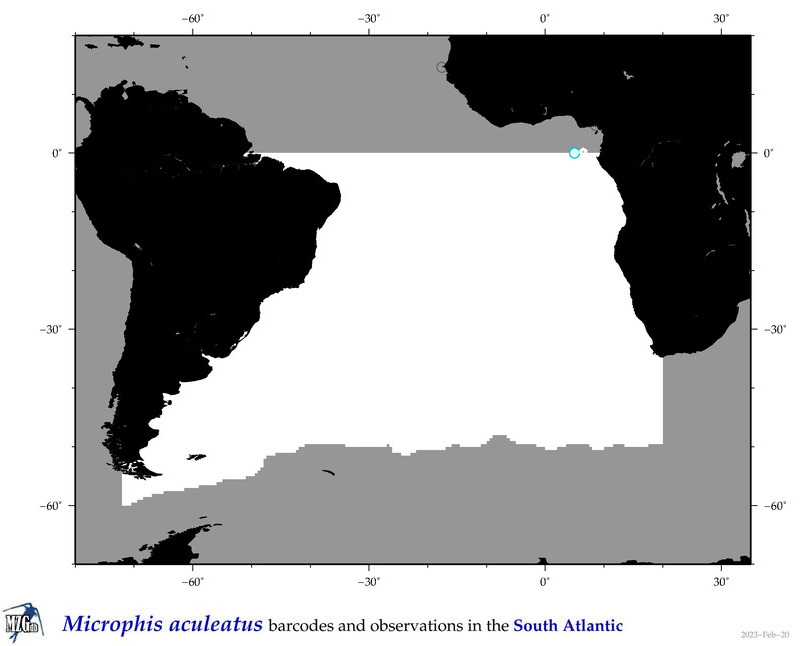
| ⋄ ⋄ ⋄ ⋄ Microphis aculeatus |
Species
(29) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 1
16S = 1
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004678
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Microphis brachyurus |
Species
(30) |
COI
12S
16S
18S
28S
|
COI = 106
12S = 8
16S = 4
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022195
R:1:1:1:0 |
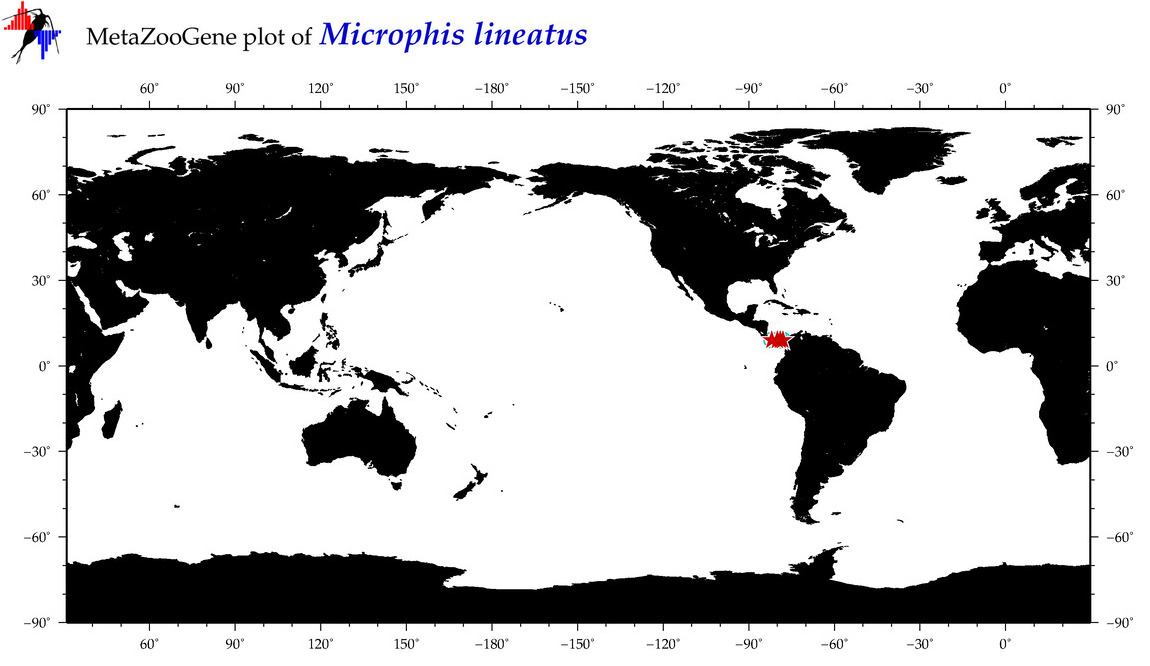
| ⋄ ⋄ ⋄ ⋄ Microphis lineatus |
Species
(31) |
COI
12S
16S
18S
28S
|
COI = 8
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004681
R:1:1:1:0 |
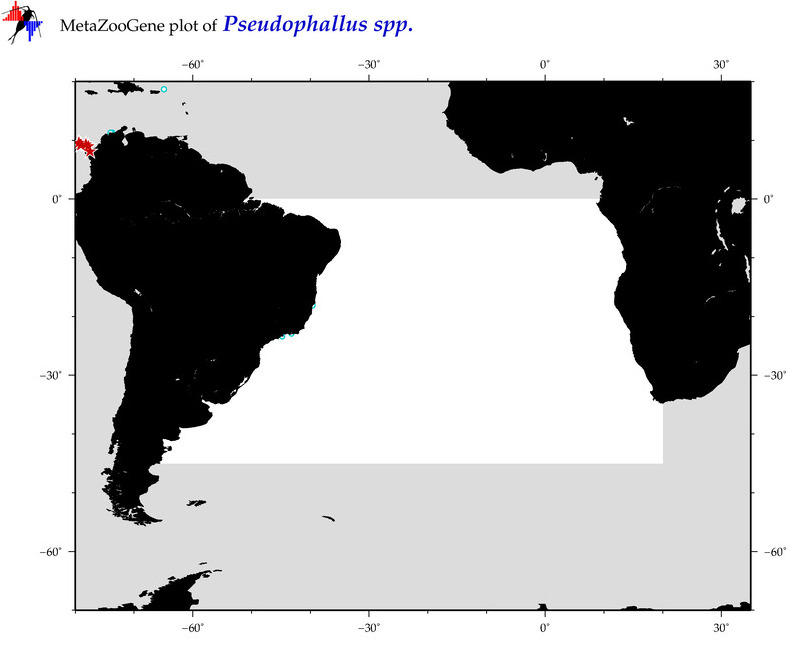
| ⋄ ⋄ ⋄ Pseudophallus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 6 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5002883
R:0:1:1:0 |
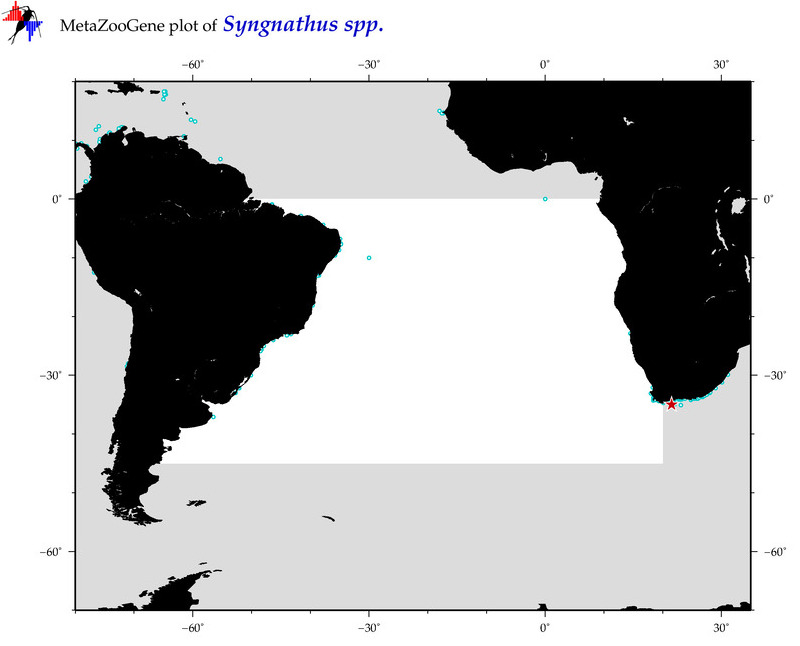
| ⋄ ⋄ ⋄ Syngnathus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 5
Sp. w/COI-Barcodes: 4
Total COI-Barcodes: 91 |
Total Species: 5
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5001322
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus acus |
Species
(32) |
COI
12S
16S
18S
28S
|
COI = 42
12S = 13
16S = 10
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005673
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus folletti |
Species
(33) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5026069
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus fuscus |
Species
(34) |
COI
12S
16S
18S
28S
|
COI = 18
12S = 3
16S = 2
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005679
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus louisianae |
Species
(35) |
COI
12S
16S
18S
28S
|
COI = 18
12S = 3
16S = 3
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
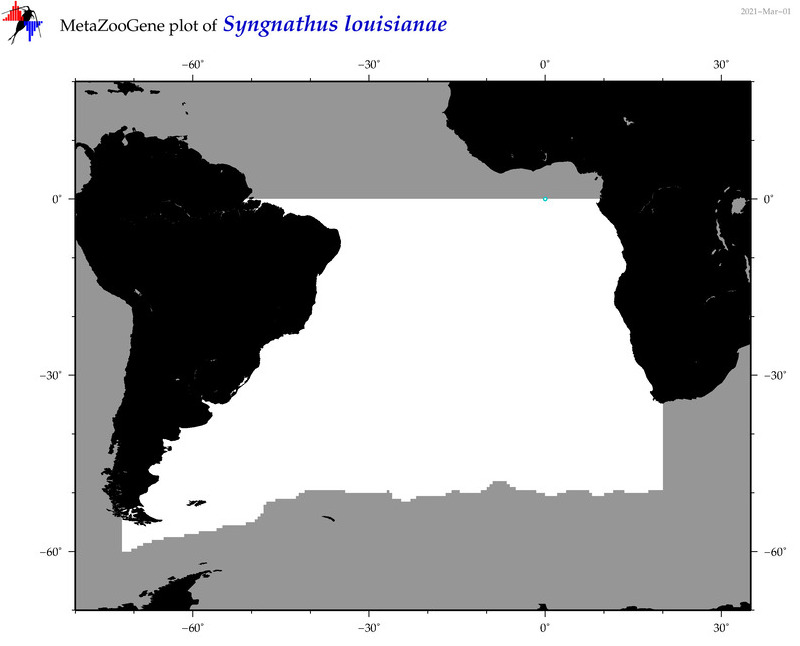 |
accepted
T5001325
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus scovelli |
Species
(36) |
COI
12S
16S
18S
28S
|
COI = 13
12S = 4
16S = 4
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
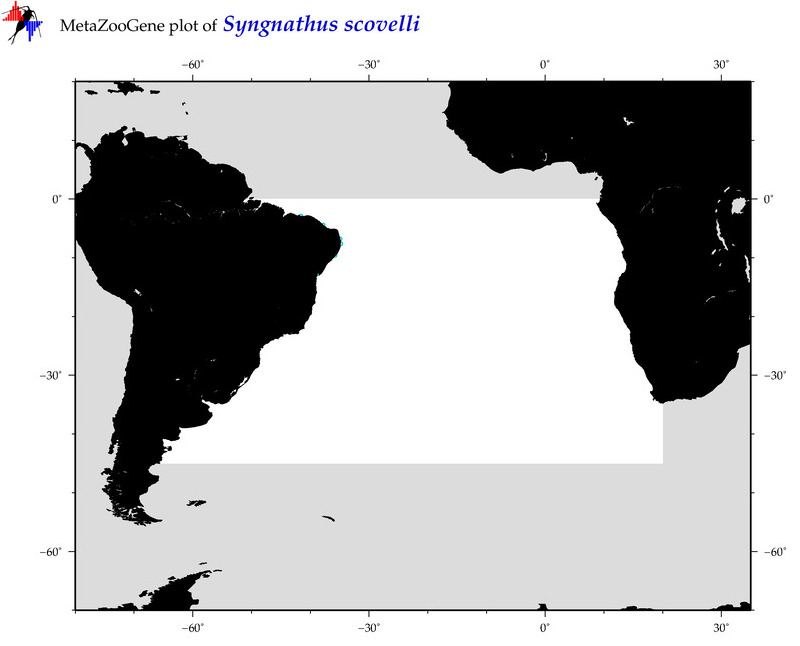 |
accepted
T5005682
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Trachyrhamphus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 5 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
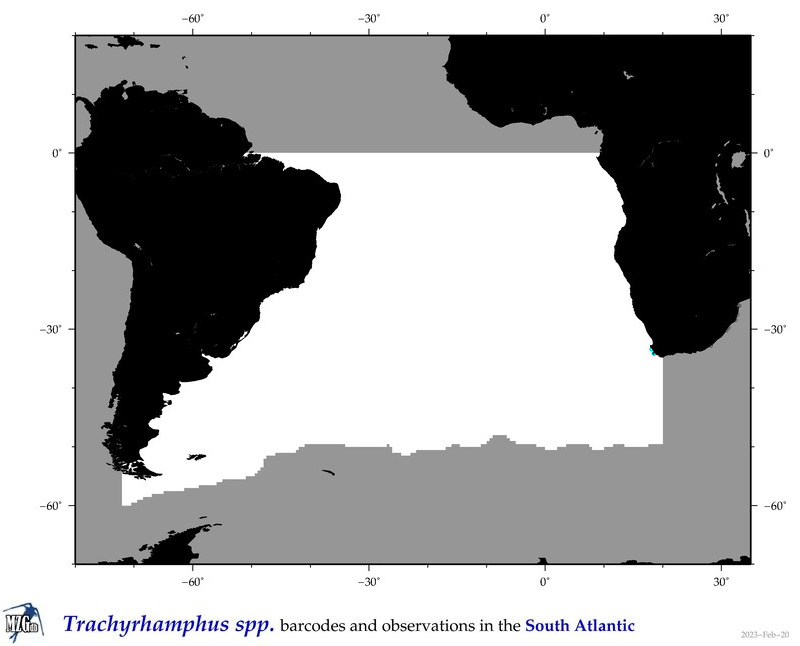 |
accepted
T5002983
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Trachyrhamphus bicoarctatus |
Species
(37) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 1
16S = 1
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
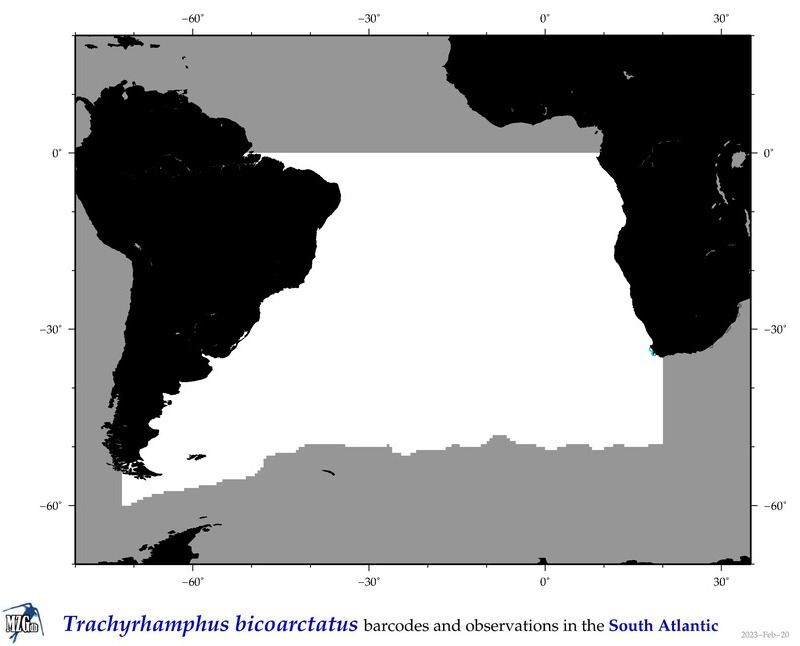 |
accepted
T5005786
R:1:0:0:0 |